Alan
Elite member
- Messages
- 2,517
- Reaction score
- 450
- Points
- 0
- Ethnic group
- Kurdish
- Y-DNA haplogroup
- R1a1a1
- mtDNA haplogroup
- HV2a1 +G13708A
Interesting comments. Here are the Eurogenes15, Dodecad12b, and DodecadV3 results for Ust' Ishim, Kostenki14 and Mal'ta:
Eurogenes15 Ust'-Ishim Kostenki
14Mal'ta Dodecad K12b Ust'-Ishim Kostenki
14Mal'ta Dodecad V3 Ust'-Ishim Kostenki
14Mal'ta North_Sea - 18.81% 15.91% Gedrosia 9.58% 12.38% 24.39% East_European 4.55% 11.89% 20.03% Atlantic 11.24% 12.39% - Siberian 2.70% 3.78% 13.55% West_European 8.95% 30.62% 37.68% Baltic - 6.52% 6.54% Northwest_African 2.58% 1.65% - Mediterranean 4.94% 15.55% - Eastern_Euro 3.08% 9.71% 38.02% Southeast_Asian 13.76% 6.12% - Neo_African 3.76% 1.43% 0.38% West_Med 4.82% 9.77% - Atlantic_Med 6.82% 21.46% - West_Asian - 0.63% - West_Asian - - - North_European 7.39% 28.80% 47.46% South_Asian 30.85% 17.75% 26.04% East_Med - - - South_Asian 31.50% 15.70% 14.36% Northeast_Asian 5.96% 4.58% 15.53% Red_Sea 3.36% 5.70% - East_African 8.24% 3.87% - Southeast_Asian 21.76% 6.72% - South_Asian 30.76% 17.42% 20.31% Southwest_Asian 3.36% 4.95% - East_African 7.03% 1.92% - Southeast_Asian 15.25% 1.33% - East_Asian 8.40% 0.15% - Southwest_Asian 3.86% 4.61% - Siberian 2.02% 0.66% - Caucasus - - - Northwest_African 3.25% 2.07% - Amerindian 2.20% 4.74% 18.62% Sub_Saharan 5.67% 1.15% 0.24% Palaeo_African 5.10% 2.24% 0.34% Oceanian 10.96% 5.11% 0.12% Northeast_African 10.08% 5.19% - Sub-Saharan 6.22% 2.66% 0.47%
None of them have the K12b Caucasus component, or the Eurogenes15 West_Asian or East_Med components, and Kostenki14 has just 0.63% of the West_Asian component in Dodecad V3.
My first thought when seeing the Eurogenes15 results for Kostenki14 was that it was contamination, but now seeing how they all lack that Caucasus / Near Eastern component makes me think it is the real deal. Why it's missing that area, and seeing how Kostenki14 is so close to there geographically though is intriguing.
The explanatio n is simple, "Gedrosia" is more of the "West Asian" component than "Caucasus" is.
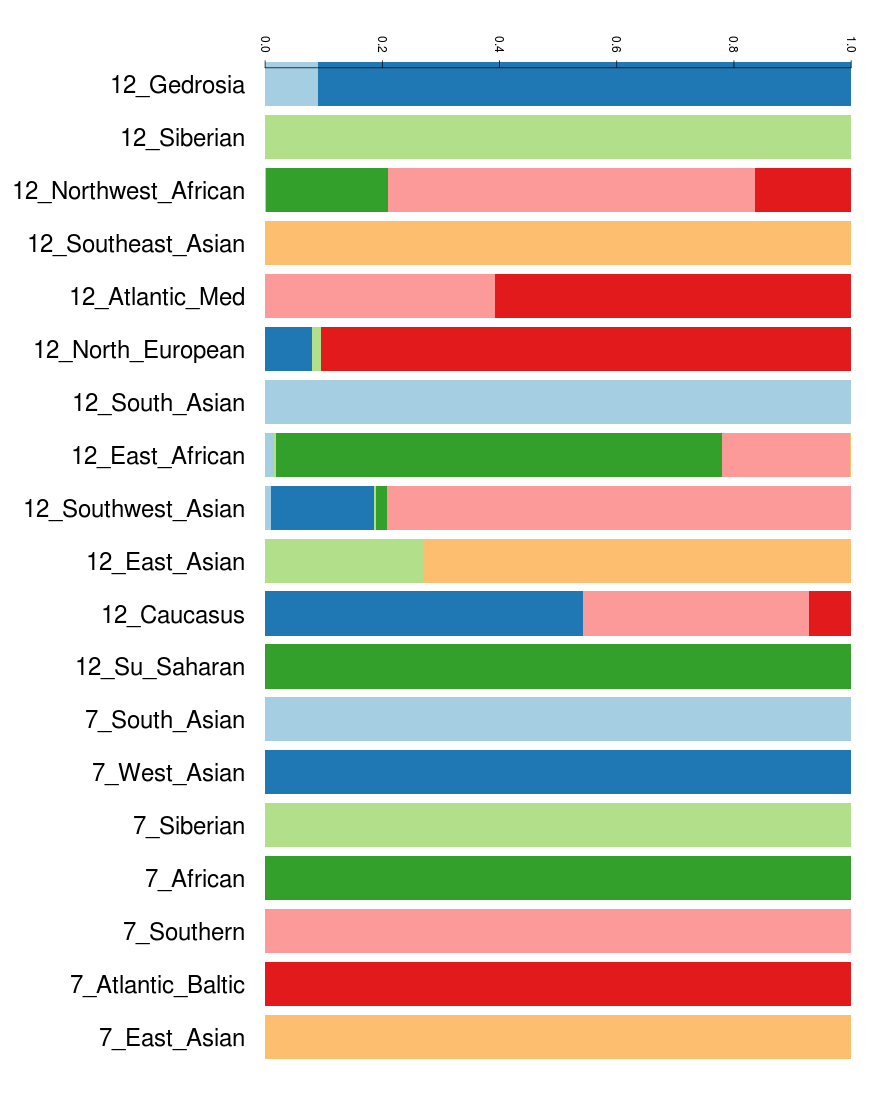
Gedrosia is 90% "West Asian" of K7b + 10% "ANI".
Caucasus is ~55% "West Asian", *37% "Southern" and ~8% Atlantic_Baltic. This is why Ötzi showed in some calculators NO West Asian but in K12b some "Caucasus". It is the "Southern" portion inside Caucasus which shows up and not the West Asian portion but for some kind of reason people seem to never understand that. The West Asian portion of Mal'ta is much more of the Gedrosia type, which makes quite sense since Gedrosia is the "purer" portion of "West Asian" and which is much more widespred in South_Central and East Asia compared to Caucasus.
Also as I wrote earlier. In Eurogenes 15 and Dodecad v3 the West Asian (Gedrosia) gets eaten up by South Asian and some of the other West Asian portions get eaten up by various "North European" components.
We know that from the Lazaridis paper where North Caucasians who score high in ANE are shown with almost 40% (North)"European" ( Most Caucasians have 20-26% North European the rest must be Caucasus which has been labeled under European) and substantial South Asian (which they in reality lack and can be only explained with that Gedrosia has been put under the South Asian label).
Davidski made it's own run with a differen't calculator on Mal'ta genome. And he came to the conclusion that the sample can be explained as 30% North European like, 25% Caucasus_Gedrosia(more Gedrosia than Caucasus) like, 25% Amerindian like and the rest was ASI and Southeast Asian like.
Last edited: