Hauteville
Elite member
- Messages
- 820
- Reaction score
- 162
- Points
- 0
- Ethnic group
- Italian
- Y-DNA haplogroup
- I-S185
- mtDNA haplogroup
- U5b2b
Gela was also a Dorian colony.
Follow along with the video below to see how to install our site as a web app on your home screen.

Note: This feature currently requires accessing the site using the built-in Safari browser.
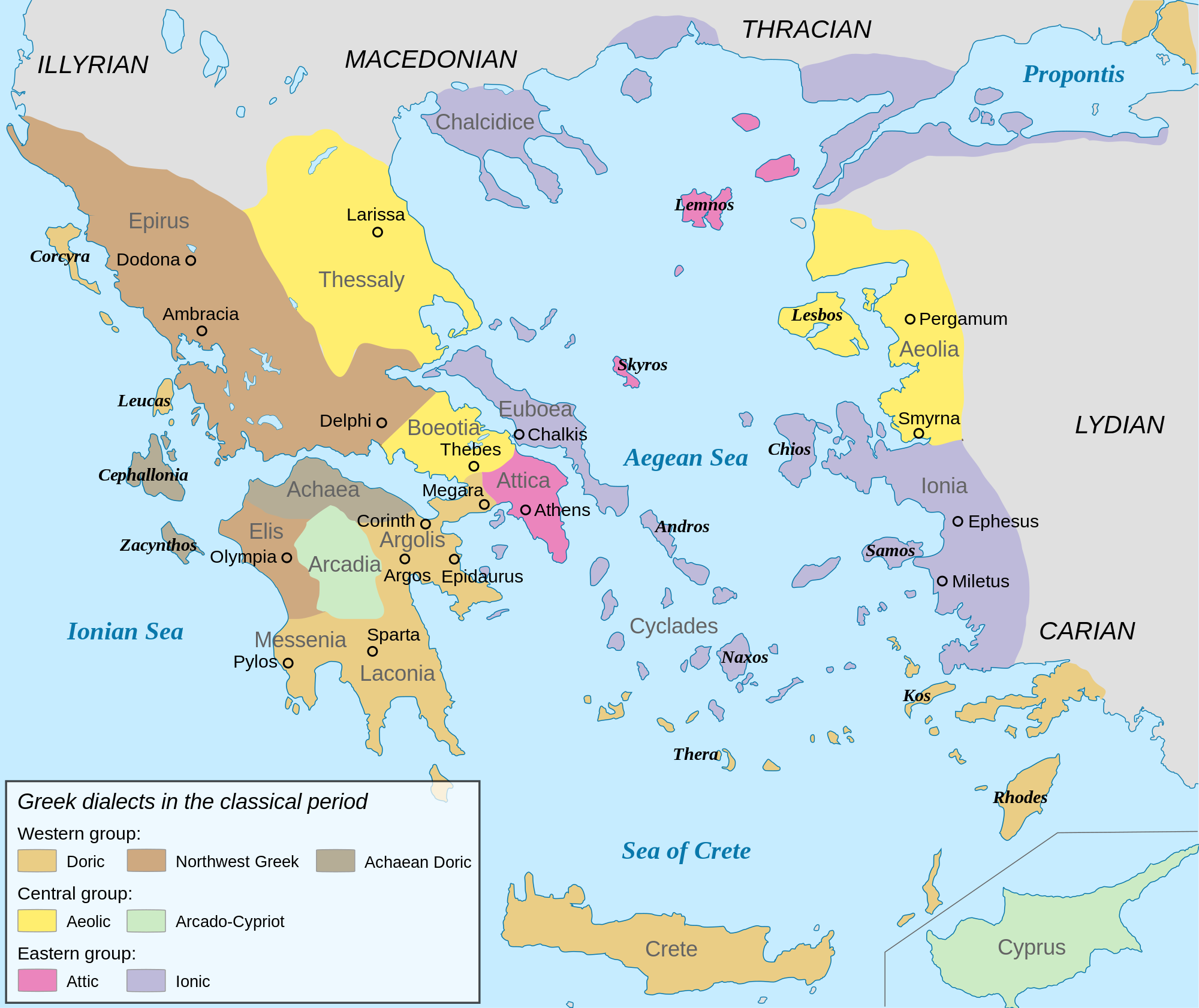
Y Dna "I" in West Sicily, with a total sample of 68:
M223 (5)
M253 (2)
M26 (4)
M423 (1)
M438 (1)
Of course, M26 is the "Sardinian" lineage.
Eastern Sicilian samples are 256, which is more interesting.I am surprised that this new study still uses very old-dated haplogroup names like E3b (dropped by ISOGG in 2008 in favour of E1b1b) or K* instead of T (listed by ISOGG since 2008). The sample sizes are very small of each region, so the percentages are not very indicative. I will integrate the data into my Y-DNA table for Italian regions and for Greece. At least it won't be totally useless.
The more samples the better, of course, but how big are the samples for individual towns in any country in most of these studies or even countries themselves? I see that our Eupedia yDna table for all of England is based on 250-500 samples? Posters here spent days discussing the results from Boattini et al 2013, and they had 984 samples for all of peninsular Italy plus the islands, and only 45 samples for Agrigento. Here we have 556 samples for southern Italy alone, 68 for Trapani, and 256 for Siracusa and a few small nearby areas. That seems pretty darn good to me, although I'd love it if Italian researchers had the money to do a POBI type study for Italy. Meanwhile, I'll take what I can get.
As for why the dated nomenclature, I can't be sure until I see the study itself. I'm wondering if these are new samples. See Table 2.
Angela was all the combined R1b in eastern Sicily 36%?
About I, the combined I in West Sicily is around 18-19% right?I'm without pc in this moment so I don't remember very well the right percentages.

This thread has been viewed 22597 times.
