sparkey
Great Adventurer
- Messages
- 2,250
- Reaction score
- 352
- Points
- 0
- Location
- California
- Ethnic group
- 3/4 Colonial American, 1/8 Cornish, 1/8 Welsh
- Y-DNA haplogroup
- I2c1 PF3892+ (Swiss)
- mtDNA haplogroup
- U4a (Cornish)
Eurogenes has recently explored a three-component admixture analysis based on ancient samples, in which the three components are named WHG (West European Hunter-Gatherer), EEF (Early European Farmer), and ANE (Ancient North Eurasian). This analysis can be applied to existing European populations. See here.
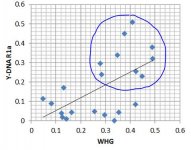
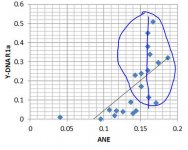
Based on existing ancient samples and analysis of modern samples, there's a strong indication that Haplogroup I was important in the source population(s) of WHG, and Haplogroup R (perhaps especially R1a?) was important in the source population(s) of ANE. But do these patterns still hold? Taking the breakdown from Eurogenes and the Y-DNA frequencies from Eupedia, I get the following scatter plots and basic regression lines:

Furthermore, we can see some interesting patterns when looking at the difference between haplogroup and component frequency population-by-population:

What to take from this? Here are some of my takes:
Based on existing ancient samples and analysis of modern samples, there's a strong indication that Haplogroup I was important in the source population(s) of WHG, and Haplogroup R (perhaps especially R1a?) was important in the source population(s) of ANE. But do these patterns still hold? Taking the breakdown from Eurogenes and the Y-DNA frequencies from Eupedia, I get the following scatter plots and basic regression lines:

Furthermore, we can see some interesting patterns when looking at the difference between haplogroup and component frequency population-by-population:

What to take from this? Here are some of my takes:
- Haplogroup I doesn't correlate with much of anything, with its strongest positive correlation being with WHG as we may expect, but even with that one, it is a weak correlation. This could be explained by the haplogroup being widespread in Europe early, but with several later expansions into non-WHG-dominant populations.
- It's also interesting to note that if we expanded this to a world admixture analysis, beyond WHG+EEF+ANE, then there would likely be a strong correlation between WHG+EEF+ANE combined and Haplogroup I.
- Haplogroup R1a's regression fit with ANE shows a good but apparently nonlinear correlation. This could indicate that beyond a certain threshold of ANE percentage, there tends to have been expansions of the R1a within those populations.
- Haplogroup R1a actually shows a correlation with WHG as well. This is good evidence for the R1a expansion into Europe (Corded Ware etc.) picking up assimilated hunter-gatherers as it went along, but not picking up farmers so much.
- I think that the average (WHG-I) and average (ANE-R1a) are significant. The fact that ave(WHG-I) is quite positive while ave(ANE-R1a) is slightly negative is strong evidence for population absorptions magnifying the Y-lines, but not the autosomal lines, of the absorbing population at the expense of the absorbed population.
Last edited: