Aberdeen
Regular Member
- Messages
- 1,835
- Reaction score
- 380
- Points
- 0
- Ethnic group
- Scottish, English and German
- Y-DNA haplogroup
- I1
- mtDNA haplogroup
- H4
Eurogenes Blog has a link to an abstract relating to a discussion paper for an upcoming conference. The authors sequenced mtDNA but apparently not Y DNA, from a location that's part of the assumed IE homeland. The abstract reads as follows.
"Genotyping of 390,000 SNPs in more than forty 3,000-9,000 year old humans from the ancient Russian steppeDavid Reich 1 ,2, Nadin Rohland1 ,2, Swapan Mallick1 ,2, Iosif Lazaridis1, Eadaoin Harney1, Susanne Nordenfelt1, Qiaomei Fu3, Matthias Meyer3, Dorcas Brown4, David Anthony4, Nick Patterson2
1Harvard Medical School, Boston, MA, USA, 2Broad Institute of Harvard and MIT, Cambridge, MA, USA, 3Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany, 4Hartwick College, Oneonta, NY, USA
A central challenge in ancient DNA research is that for many bones that contain genuine DNA, the great majority of molecules in sequencing libraries are microbial. Thus, it has been impractical to carry out whole genome analyses of substantial numbers of ancient individuals. We report a strategy for in-solution capture of ancient DNA from approximately 390,000 single nucleotide polymorphism (SNP) targets, adapting a method of Fu et al. PNAS 2013 who enriched a 40,000 year old DNA sample for the entire chromosome 21. Of the SNPs targets, the vast majority overlap the Affymetrix Human Origins array, allowing us to compare the ancient samples to a database of more than 2,700 present-day humans from 250 groups.
We applied the SNP capture as well as mitochondrial genome enrichment to a series of 65 bones dating to between 3,000-9,000 years ago from the Samara district of Russia in the far east of Europe, a region that has been suggested to be part of the Proto-Indo-European homeland. We successfully extracted nuclear data from 10-90% of targeted SNPs for more than 40 of the samples, and for all of these samples also obtained complete mitochondrial genomes. We report three key findings:
"Genotyping of 390,000 SNPs in more than forty 3,000-9,000 year old humans from the ancient Russian steppeDavid Reich 1 ,2, Nadin Rohland1 ,2, Swapan Mallick1 ,2, Iosif Lazaridis1, Eadaoin Harney1, Susanne Nordenfelt1, Qiaomei Fu3, Matthias Meyer3, Dorcas Brown4, David Anthony4, Nick Patterson2
1Harvard Medical School, Boston, MA, USA, 2Broad Institute of Harvard and MIT, Cambridge, MA, USA, 3Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany, 4Hartwick College, Oneonta, NY, USA
A central challenge in ancient DNA research is that for many bones that contain genuine DNA, the great majority of molecules in sequencing libraries are microbial. Thus, it has been impractical to carry out whole genome analyses of substantial numbers of ancient individuals. We report a strategy for in-solution capture of ancient DNA from approximately 390,000 single nucleotide polymorphism (SNP) targets, adapting a method of Fu et al. PNAS 2013 who enriched a 40,000 year old DNA sample for the entire chromosome 21. Of the SNPs targets, the vast majority overlap the Affymetrix Human Origins array, allowing us to compare the ancient samples to a database of more than 2,700 present-day humans from 250 groups.
We applied the SNP capture as well as mitochondrial genome enrichment to a series of 65 bones dating to between 3,000-9,000 years ago from the Samara district of Russia in the far east of Europe, a region that has been suggested to be part of the Proto-Indo-European homeland. We successfully extracted nuclear data from 10-90% of targeted SNPs for more than 40 of the samples, and for all of these samples also obtained complete mitochondrial genomes. We report three key findings:
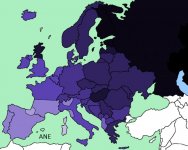
- Samples from the Samara region possess Ancient North Eurasian (ANE) admixture related to a recently published 24,000 year old Upper Paleolithic Siberian genome. This contrasts with both European agriculturalists and with European hunter-gatherers from Luxembourg and Iberia who had little such ancestry (Lazaridis et al. arXiv.org 2013). This suggests that European steppe groups may have been be implicated in the dispersal of ANE ancestry across Europe where it is currently pervasive.
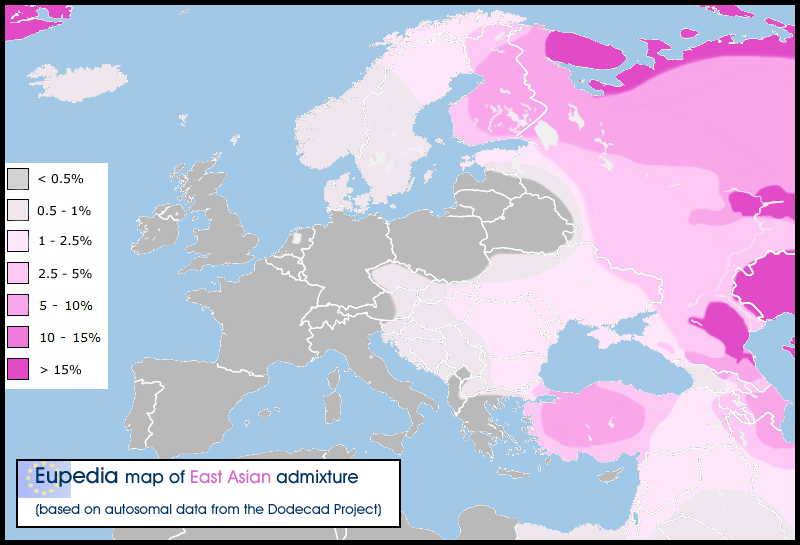
- The mtDNA composition of the steppe population is primarily West Eurasian, in contrast with northwest Russian samples of this period (Der Sarkissian et al. PLoS Genetics 2013) where an East Eurasian presence is evident.
- Samara experienced major population turnovers over time: early samples (>6000 years) belong primarily to mtDNA haplogroups U4 and U5, typical of European hunter-gatherers but later ones include haplogroups W, H, T, I, K, J.