Angela
Elite member
- Messages
- 21,823
- Reaction score
- 12,329
- Points
- 113
- Ethnic group
- Italian
I wish we had yDna, and autosomal, but still, it's interesting, and it's based on whole mitochondria.
See: A.S. Sokolov et al
"Six complete mitochondrial genomes from Early Bronze Age humans in the North Caucasus"
http://www.sciencedirect.com/science/article/pii/S0305440316301091
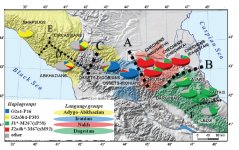
"The North Caucasus region is rich in early Bronze Age sites, with burials yielding many artifacts, including those from the Chekon, Natukhaevskaya, Katusvina-Krivitsa kurgan groups (at Krasnodar Krai, Russia) and Klady kurgan (near Novosvobodnaya Village, Republic of Adygea, Russia). According to the mainstream archaeological hypothesis, these sites belong to the Maikop culture (3700–3000 years BC), with Novosvobodnaya communities representing an offshoot of Maikop ancestry. However, due to specific differences in Novosvobodnaya artifacts, the Maikop and Novosvobodnaya assemblages could represent two synchronous archaeological cultures living in almost sympatry but showing independent ancestry, from the Near East and Europe respectively. Here, we used target-enrichment together with high-throughput sequencing to characterize the complete mitochondrial sequence of three Maikop and three Novosvobodnaya individuals. We identified T2b, N1b1 and V7 haplogroups, all widely spread in Neolithic Europe. In addition, we identified the Paleolithic Eurasian U8b1a2 and M52 haplogroups, which are frequent in modern South Asia, particularly in modern India. Our data provide a deeper understanding of the diversity of Early Bronze Age North Caucasus communities and hypotheses of its origin. Analyzing non-human sequencing reads for microbial content, we found that one individual from the Klady kurgan was infected by the pathogen Brucella abortus that is responsible for zoonotic infections from cattle to humans. This finding is in agreement with Maikop/Novosvobodnaya livestock groups, mostly consisting of domestic pigs and cattle. This paper represents a first mitochondrial genome analysis of Maikop/Novosvobodnaya culture as well as the earliest brucellosis case in archaeological humans."
The site was previously studied and they also found mtDna V7. Here is the link:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4115223/
" The mtDNA haplogroup affiliation was determined as V7, suggesting a role of the TRB culture in the development of the Novosvobodnaya culture and supporting the model of sharing between Novosvobodnaya and early Indo-European cultures."
"One hypothesizes the existence of a single Maikop culture with two developmental phases [1-3], including finds discovered in Novosvobodnaya stanitsa (former Tsarskaya). The other hypothesis suggests that the archaeological collections assembled in Novosvobodnaya stanitsa should be treated individually, as independent artifacts (as a distinct culture). During the archaeological excavations of the kurgan grave “Klady” near Novosvobodnaya stanitsa in 1979–1991, which were supervised by A.D. Rezepkin, a total of 22 kurgans were uncovered with 93 well-stratified burial sites. These records allow one not only to establish the absolute chronology of the artifacts, but also to contribute to a better understanding of the origin of the Novosvobodnaya culture [4, 5]."
"Recently, archaeological evidence has emerged to argue against the opinion that the Novosvobodnaya culture shares links with the West Asian Maikop culture. The discovered artifacts support the hypothesis that the Baalberg phase of early periods of the Indo-European Funnel-Beaker culture played a significant role in the Novosvobodnaya archaeological culture, rather than the West Asian Maikop culture [5]. To prove or rule out this hypothesis, a DNA analysis is required as one of the definitive tools."
I can't say the results are exactly what I would have expected. I would have thought there would have been some U4/U5 in this group, although this is only three samples and those may show up if we get more samples.
As to Maykop, I expected south of Caucasus mtDna, but I didn't expect mtDna so affiliated with India.
I don't see how the Maykop women marrying into the steppe could be responsible for the "CHG" there, given we don't see much of this on the steppe.
If someone has access it would be great to get the results by date and specific site at least.
As for Brucella:
"Brucellosis can affect any organ or organ system, and 90% of patients have a cyclical (undulant) fever. Though variable, symptoms can also include these clinical signs: headache, weakness, arthralgia, depression, weight loss, fatigue, and liver dysfunction. Foul-smelling perspiration is considered a classical sign. Between 20 and 60% of cases have osteoarticular complications - arthritis, spondylitis, or osteomyelitis. Hepatomegaly may occur, as can gastrointestinal complications.Up to 20% of cases can have genitourinary involvement; orchitis and epididymitis are most common. Neurological symptoms include depression and mental fatigue. Cardiovascular involvement can include endocarditis resulting in death."
https://en.wikipedia.org/wiki/Brucella
Cows may be cleaner than chickens but they still harbor lots of nasty stuff.
If the "Indo-Europeans" had some degree of immunity to this, that, along with some immunity to plague, could explain some of their success as they moved into Europe and India.
Ed.
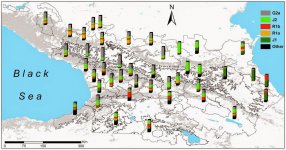
"- Krasnodar Krai, Maikop burial, 4000-3000 BCE, mt-hg U8b1a2
- Krasnodar Krai, Maikop burial, 3700-3300 BCE mt-hg U8b1a2
- Republic of Adygea, Maikop burial, Russia, 3700-3300 BCE mt-hg M52
- Republic of Adygea, Novosvobodnaya burial, Russia, 3700-3300 BCE mt-hg V7
- Krasnodar Krai, unknown burial, Russia, 3700-3300 BCE mt-hg N1b1
- Republic of Adygea, unknown burial, Russia, 3700-3300 BCE mt-hg T2b"
See: A.S. Sokolov et al
"Six complete mitochondrial genomes from Early Bronze Age humans in the North Caucasus"
http://www.sciencedirect.com/science/article/pii/S0305440316301091
"The North Caucasus region is rich in early Bronze Age sites, with burials yielding many artifacts, including those from the Chekon, Natukhaevskaya, Katusvina-Krivitsa kurgan groups (at Krasnodar Krai, Russia) and Klady kurgan (near Novosvobodnaya Village, Republic of Adygea, Russia). According to the mainstream archaeological hypothesis, these sites belong to the Maikop culture (3700–3000 years BC), with Novosvobodnaya communities representing an offshoot of Maikop ancestry. However, due to specific differences in Novosvobodnaya artifacts, the Maikop and Novosvobodnaya assemblages could represent two synchronous archaeological cultures living in almost sympatry but showing independent ancestry, from the Near East and Europe respectively. Here, we used target-enrichment together with high-throughput sequencing to characterize the complete mitochondrial sequence of three Maikop and three Novosvobodnaya individuals. We identified T2b, N1b1 and V7 haplogroups, all widely spread in Neolithic Europe. In addition, we identified the Paleolithic Eurasian U8b1a2 and M52 haplogroups, which are frequent in modern South Asia, particularly in modern India. Our data provide a deeper understanding of the diversity of Early Bronze Age North Caucasus communities and hypotheses of its origin. Analyzing non-human sequencing reads for microbial content, we found that one individual from the Klady kurgan was infected by the pathogen Brucella abortus that is responsible for zoonotic infections from cattle to humans. This finding is in agreement with Maikop/Novosvobodnaya livestock groups, mostly consisting of domestic pigs and cattle. This paper represents a first mitochondrial genome analysis of Maikop/Novosvobodnaya culture as well as the earliest brucellosis case in archaeological humans."
The site was previously studied and they also found mtDna V7. Here is the link:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4115223/
" The mtDNA haplogroup affiliation was determined as V7, suggesting a role of the TRB culture in the development of the Novosvobodnaya culture and supporting the model of sharing between Novosvobodnaya and early Indo-European cultures."
"One hypothesizes the existence of a single Maikop culture with two developmental phases [1-3], including finds discovered in Novosvobodnaya stanitsa (former Tsarskaya). The other hypothesis suggests that the archaeological collections assembled in Novosvobodnaya stanitsa should be treated individually, as independent artifacts (as a distinct culture). During the archaeological excavations of the kurgan grave “Klady” near Novosvobodnaya stanitsa in 1979–1991, which were supervised by A.D. Rezepkin, a total of 22 kurgans were uncovered with 93 well-stratified burial sites. These records allow one not only to establish the absolute chronology of the artifacts, but also to contribute to a better understanding of the origin of the Novosvobodnaya culture [4, 5]."
"Recently, archaeological evidence has emerged to argue against the opinion that the Novosvobodnaya culture shares links with the West Asian Maikop culture. The discovered artifacts support the hypothesis that the Baalberg phase of early periods of the Indo-European Funnel-Beaker culture played a significant role in the Novosvobodnaya archaeological culture, rather than the West Asian Maikop culture [5]. To prove or rule out this hypothesis, a DNA analysis is required as one of the definitive tools."
I can't say the results are exactly what I would have expected. I would have thought there would have been some U4/U5 in this group, although this is only three samples and those may show up if we get more samples.
As to Maykop, I expected south of Caucasus mtDna, but I didn't expect mtDna so affiliated with India.
I don't see how the Maykop women marrying into the steppe could be responsible for the "CHG" there, given we don't see much of this on the steppe.
If someone has access it would be great to get the results by date and specific site at least.
As for Brucella:
"Brucellosis can affect any organ or organ system, and 90% of patients have a cyclical (undulant) fever. Though variable, symptoms can also include these clinical signs: headache, weakness, arthralgia, depression, weight loss, fatigue, and liver dysfunction. Foul-smelling perspiration is considered a classical sign. Between 20 and 60% of cases have osteoarticular complications - arthritis, spondylitis, or osteomyelitis. Hepatomegaly may occur, as can gastrointestinal complications.Up to 20% of cases can have genitourinary involvement; orchitis and epididymitis are most common. Neurological symptoms include depression and mental fatigue. Cardiovascular involvement can include endocarditis resulting in death."
https://en.wikipedia.org/wiki/Brucella
Cows may be cleaner than chickens but they still harbor lots of nasty stuff.
If the "Indo-Europeans" had some degree of immunity to this, that, along with some immunity to plague, could explain some of their success as they moved into Europe and India.
Ed.
"- Krasnodar Krai, Maikop burial, 4000-3000 BCE, mt-hg U8b1a2
- Krasnodar Krai, Maikop burial, 3700-3300 BCE mt-hg U8b1a2
- Republic of Adygea, Maikop burial, Russia, 3700-3300 BCE mt-hg M52
- Republic of Adygea, Novosvobodnaya burial, Russia, 3700-3300 BCE mt-hg V7
- Krasnodar Krai, unknown burial, Russia, 3700-3300 BCE mt-hg N1b1
- Republic of Adygea, unknown burial, Russia, 3700-3300 BCE mt-hg T2b"