Pax Augusta
Elite member
A study has been published recently that analyzed the mitochondrial DNA of 30 samples from an Iron Age archaeological site in Sicily associated with the Sicani people.
New Insights Into Mitochondrial DNA Reconstruction and Variant Detection in Ancient Samples
Front. Genet., 18 February 2021 | https://doi.org/10.3389/fgene.2021.619950
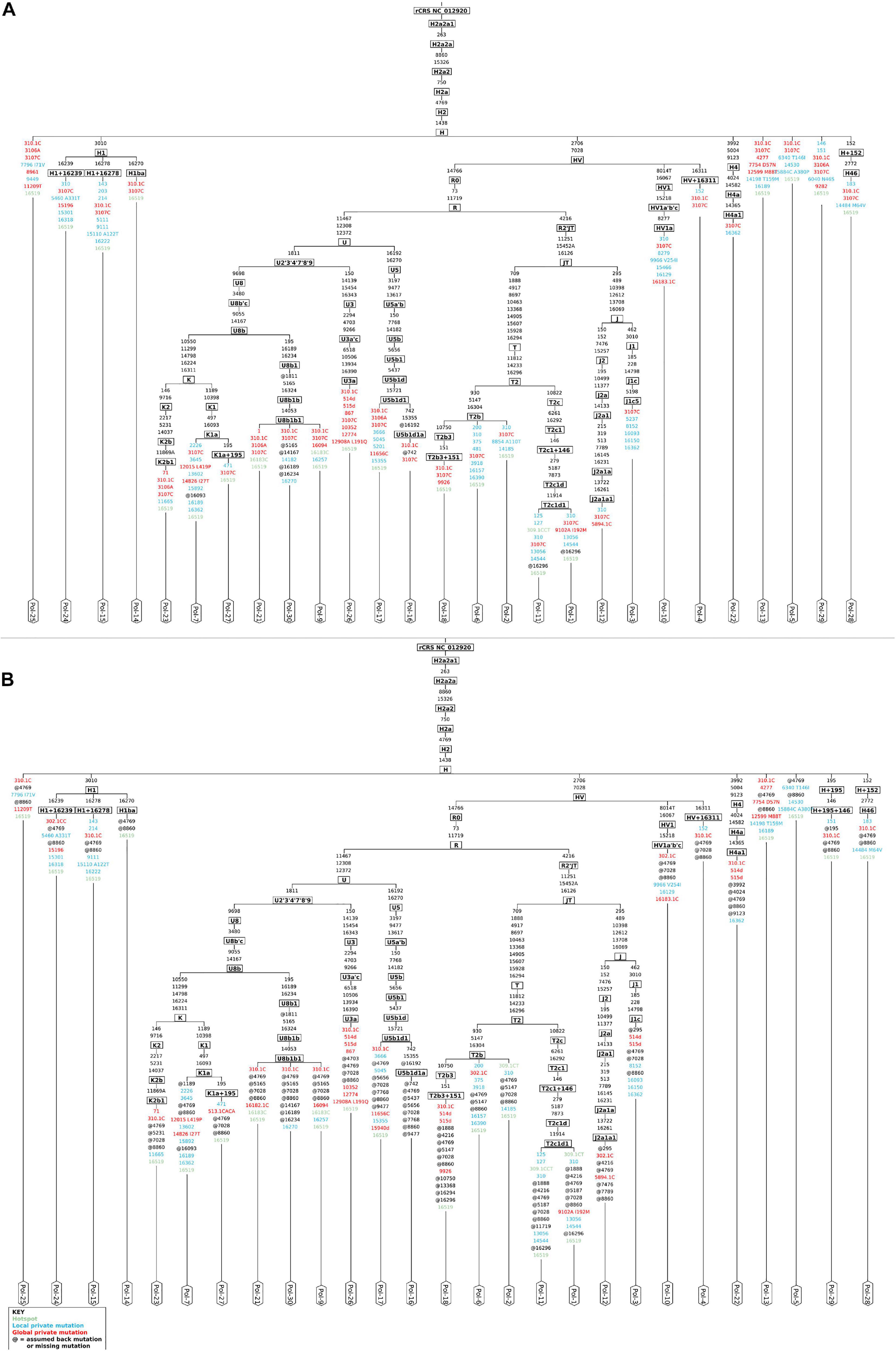
"We implemented and applied a computational pipeline for mtDNA analysis to a dataset of 30 ancient human samples from an Iron Age necropolis in Polizzello (Sicily, Italy). The pipeline includes several modules from well-established tools for aDNA analysis and a recently released variant caller, which was specifically conceived for mtDNA, applied for the first time to aDNA data. Through a fine-tuned filtering on variant allele sequencing features, we were able to accurately reconstruct nearly complete (>88%) mtDNA genome for almost all the analyzed samples (27 out of 30), depending on the degree of preservation and the sequencing throughput, and to get a reliable set of variants allowing haplogroup prediction."
"we analyzed 30 Iron Age samples from Polizzello (Sicily, Italy), an archeological site in the heart of Sikania dated at IX–VII centuries BC (Messina et al., 2008)."
"The good quality of endogenous mitochondrial consensus sequences assembled was confirmed by an accurate mitochondrial haplogroup prediction, involving R0, JT, and U subtrees, all descending from the R clade (Phylotree Build 17), quite in line with their European origin. "
https://www.frontiersin.org/articles/10.3389/fgene.2021.619950/full
Polizzello archaeological site (wikipedia)
https://en.wikipedia.org/wiki/Polizzello_archaeological_site

New Insights Into Mitochondrial DNA Reconstruction and Variant Detection in Ancient Samples
Front. Genet., 18 February 2021 | https://doi.org/10.3389/fgene.2021.619950
"We implemented and applied a computational pipeline for mtDNA analysis to a dataset of 30 ancient human samples from an Iron Age necropolis in Polizzello (Sicily, Italy). The pipeline includes several modules from well-established tools for aDNA analysis and a recently released variant caller, which was specifically conceived for mtDNA, applied for the first time to aDNA data. Through a fine-tuned filtering on variant allele sequencing features, we were able to accurately reconstruct nearly complete (>88%) mtDNA genome for almost all the analyzed samples (27 out of 30), depending on the degree of preservation and the sequencing throughput, and to get a reliable set of variants allowing haplogroup prediction."
"we analyzed 30 Iron Age samples from Polizzello (Sicily, Italy), an archeological site in the heart of Sikania dated at IX–VII centuries BC (Messina et al., 2008)."
"The good quality of endogenous mitochondrial consensus sequences assembled was confirmed by an accurate mitochondrial haplogroup prediction, involving R0, JT, and U subtrees, all descending from the R clade (Phylotree Build 17), quite in line with their European origin. "
https://www.frontiersin.org/articles/10.3389/fgene.2021.619950/full
Polizzello archaeological site (wikipedia)
https://en.wikipedia.org/wiki/Polizzello_archaeological_site