Philjames100
Regular Member
- Messages
- 309
- Reaction score
- 130
- Points
- 43
More info on lactase persistence in Africa:
Sahelian pastoralism from the perspective of variants associated with lactase persistence (Priehodová et al. 2020)
Abstract
Objectives: Archeological evidence shows that first nomadic pastoralists came to the African Sahel from northeastern Sahara, where milking is reported by 7.5 ka. A second wave of pastoralists arrived with the expansion of Arabic tribes in 7th–14th century CE. All Sahelian pastoralists depend on milk production but genetic diversity underlying their lactase persistence (LP) is poorly understood. Materials and methods: We investigated SNP variants associated with LP in 1,241 individuals from 29 mostly pastoralist populations in the Sahel. Then, we analyzed six SNPs in the neighboring fragment (419 kb) in the Fulani and Tuareg with the −13910*T mutation, reconstructed haplotypes, and calculated expansion age and growth rate of this variant.
Results: Our results reveal a geographic localization of two different LP variants in the Sahel: −13910*T west of Lake Chad (Fulani and Tuareg pastoralists) and −13915*G east of there (mostly Arabic-speaking pastoralists). We show that −13910*T has a more diversified haplotype background among the Fulani than among the Tuareg and that the age estimate for expansion of this variant among the Fulani (8.5 ka) corresponds to introduction of cattle to the area.
Conclusions: This is the first study showing that the “Eurasian” LP allele −13910*T is widespread both in northern Europe and in the Sahel; however, it is limited to pastoralists in the Sahel. Since the Fulani haplotype with −13910*T is shared with contemporary Eurasians, its origin could be in a region encompassing the Near East and northeastern Africa in a population ancestral to both Saharan pastoralists and European farmers.
Link


Excerpts:
Although the auroch, ancestor of the domestic cattle, also lived in northeastern Africa, as evidenced by rock art and archeological findings, its domestication took place in Asia. The auroch was domesticated independently in the Near East and in India and two different breeds were subsequently introduced to Africa. (...) In sub-Saharan Africa and Arabia, the first food-producing societies were nomadic pastoralists. (...)
Several variants associated with LP are found in current populations. There are three different mutations in eastern Africa (−14010*C, −14009*G, and −13907*G), one in western Eurasia and northwestern Africa (−13910*T), and one in Arabia and northeastern Africa (−13915*G). These mutations can be viewed as evidence of convergent genetic adaptation of humans to pastoralism. It has been estimated that allele −13910*T is the oldest among these variants. Simulation models show that it emerged somewhere between central Europe and northern Balkans at 7.5 ka, but its presence has not yet been attested by ancient DNA studies of European human remains from this period. Currently the prevailing view is that this allele had been present in the population at a very low frequency and became strongly selected for much later, perhaps during the Bronze Age, in conjunction with other alleles associated with lipid metabolism.
As mentioned above, the −13910*T variant occurs not only in western Eurasia but also in northwestern Africa. Based on high frequencies of −13910*T and Eurasian Y chromosome haplotypes in the Fulani (Fulbe) from Cameroon, it has been suggested that this LP variant may have been introduced in their population from outside Africa. Based on (probably different) Fulani DNA samples but again from northern Cameroon, it has been demonstrated that their −13910*T variant shares its haplotype backgrounds with Europeans, which suggests a single mutation event for this mutation and its introduction via admixture of non-African sources and through gene flow. A recent study of Fulani pastoralists from Ziniaré, Burkina Faso (Vicente et al., 2019) confirmed these suggestions and showed that this variant shares its haplotype background with Eurasians and that ancestors of the Fulani probably acquired the haplotype via recent admixture events which took place in northern Africa. (...)
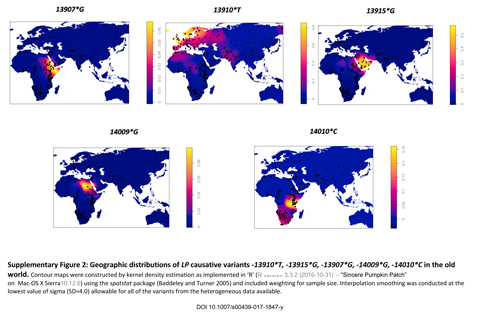
We screened the sequences for five variants (−13907*G, −13910*T, −13915*G, −14009*G, and −14010*C) associated with LP. The most frequent variant in our dataset, −13910*T, was found in all analyzed western Sahelian pastoralists such as the Fulani, Tuareg, and Moors in more or less similar frequencies but was not present in other groups. The second most widespread variant in our dataset, −13915*G, was found in seven populations living in the eastern part of the region. It occurred at the highest frequency among the Kababish Arabs (33.3%), while in other populations, its frequency was rather low. The third most frequent variant was −14009*G, found almost exclusively among the Beja (42.6%): in other groups, it was present at much lower frequencies. Variant −13907*G was found in seven Afar, five Beja, and one Kababish individual, and variant −14010*C was not found in our dataset at all. The LP variants of the Sahelian and neighboring populations are shown in Figure 1. (...)
Using the method of Austerlitz et al. (2003), we estimated the expansion age of −13910*T in the Fulani population at 7,534–9,686 ya, along with a slow growth rate of 1.033–1.053. (...)
Discussion
Our investigation of LP variant frequencies revealed new and interesting results related to the origins of pastoralism and subsequent gene flow between pastoralists and farmers in the Sahel/Savannah belt of Africa. We observed a clear distinction between regions west and east of Lake Chad: while variant −13910*T prevails in the western Sahel, where we found it only in pastoralists such as the Fulani, Tuareg, and Moors (it is virtually absent in sedentary farmers analyzed in our dataset), eastern Sahel appears to be much more diverse and several LP-associated variants, such as −13907*G, −13915*G, and 14010*C, are found in various populations.
The −14009*G variant has been associated with LP in eastern Africa (Jones et al., 2013; Jones et al., 2015) and frequency at which we found it in the Beja near Kassala, Sudan (42.6%) is much higher than reported by other studies. (...)
Our study shows that the age of expansion of −13910*T obtained for the Fulani pastoralists in western Sahel/Savannah belt (7,534–9,686 years ago) is much older than that obtained for the −13915*G in Arabic pastoralists in eastern Sahel/Savannah belt, when applying the same method. Priehodová et al., 2017 estimated age of −13915*G at between 1,274 and 1,782 years ago, which is consistent with the time of Arabic expansion to Africa. The estimate obtained in this study for the Fulani pastoralists conforms to estimates for introduction of pastoralism in Africa which are based on archeological evidence. (...)
This first Saharan/Sahelian influx of population with LP individuals was followed by a later migration of other pastoralists from the Arabian Peninsula throughout northern Africa and Sahara up to the Lake Chad Basin, which was led by Bedouin Arabic-speaking tribes. This migration probably introduced the −13915*G variant to the region, which originated in Arabia as an adaptation to the consumption of camel milk. (...)
It has been suggested that the Fulani acquired the −13910*T haplotype by admixing with a North African population which had some Eurasian ancestry, so that this variant in the Fulani population has the same descent as the variant found in Eurasia. Furthermore, it has been estimated that two distinct admixture events took place, one 1,828 and the other 302 years ago (Vicente et al., 2019). Our estimates of the growth and selection of −13910*T haplotype in the Fulani indicate an earlier dating than the above mentioned admixture times (Vicente et al., 2019) and are consistent rather with the estimates of Coelho et al. (2005), who proposes a period of 10,125–6,060 years ago.
The age of the second main LP Sahelian variant −13915*G, which we estimated at 1.5 ka, perfectly matches the timing of migration of Bedouin tribes from Arabia to Africa. A previous estimate of the age of −13915*G, which was up to 4 ka (Enattah et al., 2008), concerns its origin in Arabia, probably Central Arabia. From there, it expanded at the beginning of Middle Ages, therefore much later than the −13910*T variant, which was already established in the western Sahelian populations.
This discrepancy between the age estimates of −13910*T in the Fulani population can be explained by the fact that the two studies, that is, our present study and Vicente et al.'s (2019), targeted different parts of the genome (a local haplotype age of growth estimate vs. a genome-wide estimate of admixture), used different dating methods, and the samples on which they were based covered different populations. Moreover, Vicente et al. (2019) analyzed only one Fulani population (from Ziniaré, Burkina Faso), while our results are based on a larger Fulani dataset stretching geographically from Senegal to Chad. Nevertheless, we want to stress that our results do not exclude the possibility of a later admixture and introgression as suggested by Vicente et al., 2019. In fact, the TAS2R genes alleles may have also been introduced to the Fulani via recent admixture with a Eurasian population (Triska et al., 2015). In short, these results reflect the complexity of the population history of Africa. (...)
Because the −13910*T haplotype associated with LP is shared between the Fulani from Ziniaré, Burkina Faso, and Europeans (Vicente et al., 2019) and since we found an old age of expansion for this mutation within the African population of the Fulani (dataset covering much larger area of the Sahel), it is possible that the −13910*T may have originated in a food-producing population that lived in the Near East, including northeastern Africa, as suggested by Myles et al. (2005). Subsequently, though, the first Neolithic farmers who lived in the Near East/northeastern Africa were replaced by various population movements (Černý, Čížková, Poloni, Al-Meeri, & Mulligan, 2016; Vyas et al., 2016), with the result that only the originally “Arabian” –13,915*G LP variant is present in this region today (Priehodová et al., 2014). (...)
Interestingly, genetically the nearest population to the first Near Eastern farmers has been conserved in Sardinia (Chiang et al., 2018; Sikora et al., 2014), where the frequency of −13910*T is as low as elsewhere in the Mediterranean (Gerbault, 2013; Meloni et al., 2001). (...)
later migrations between Arabia, Near East, and North Africa (Černý et al., 2016; Fernandes et al., 2015) may be responsible for the fact that variant −13910*T is almost entirely absent from the Near East today (Gerbault et al., 2011)"
Sahelian pastoralism from the perspective of variants associated with lactase persistence (Priehodová et al. 2020)
Abstract
Objectives: Archeological evidence shows that first nomadic pastoralists came to the African Sahel from northeastern Sahara, where milking is reported by 7.5 ka. A second wave of pastoralists arrived with the expansion of Arabic tribes in 7th–14th century CE. All Sahelian pastoralists depend on milk production but genetic diversity underlying their lactase persistence (LP) is poorly understood. Materials and methods: We investigated SNP variants associated with LP in 1,241 individuals from 29 mostly pastoralist populations in the Sahel. Then, we analyzed six SNPs in the neighboring fragment (419 kb) in the Fulani and Tuareg with the −13910*T mutation, reconstructed haplotypes, and calculated expansion age and growth rate of this variant.
Results: Our results reveal a geographic localization of two different LP variants in the Sahel: −13910*T west of Lake Chad (Fulani and Tuareg pastoralists) and −13915*G east of there (mostly Arabic-speaking pastoralists). We show that −13910*T has a more diversified haplotype background among the Fulani than among the Tuareg and that the age estimate for expansion of this variant among the Fulani (8.5 ka) corresponds to introduction of cattle to the area.
Conclusions: This is the first study showing that the “Eurasian” LP allele −13910*T is widespread both in northern Europe and in the Sahel; however, it is limited to pastoralists in the Sahel. Since the Fulani haplotype with −13910*T is shared with contemporary Eurasians, its origin could be in a region encompassing the Near East and northeastern Africa in a population ancestral to both Saharan pastoralists and European farmers.
Link


Excerpts:
Although the auroch, ancestor of the domestic cattle, also lived in northeastern Africa, as evidenced by rock art and archeological findings, its domestication took place in Asia. The auroch was domesticated independently in the Near East and in India and two different breeds were subsequently introduced to Africa. (...) In sub-Saharan Africa and Arabia, the first food-producing societies were nomadic pastoralists. (...)
Several variants associated with LP are found in current populations. There are three different mutations in eastern Africa (−14010*C, −14009*G, and −13907*G), one in western Eurasia and northwestern Africa (−13910*T), and one in Arabia and northeastern Africa (−13915*G). These mutations can be viewed as evidence of convergent genetic adaptation of humans to pastoralism. It has been estimated that allele −13910*T is the oldest among these variants. Simulation models show that it emerged somewhere between central Europe and northern Balkans at 7.5 ka, but its presence has not yet been attested by ancient DNA studies of European human remains from this period. Currently the prevailing view is that this allele had been present in the population at a very low frequency and became strongly selected for much later, perhaps during the Bronze Age, in conjunction with other alleles associated with lipid metabolism.
As mentioned above, the −13910*T variant occurs not only in western Eurasia but also in northwestern Africa. Based on high frequencies of −13910*T and Eurasian Y chromosome haplotypes in the Fulani (Fulbe) from Cameroon, it has been suggested that this LP variant may have been introduced in their population from outside Africa. Based on (probably different) Fulani DNA samples but again from northern Cameroon, it has been demonstrated that their −13910*T variant shares its haplotype backgrounds with Europeans, which suggests a single mutation event for this mutation and its introduction via admixture of non-African sources and through gene flow. A recent study of Fulani pastoralists from Ziniaré, Burkina Faso (Vicente et al., 2019) confirmed these suggestions and showed that this variant shares its haplotype background with Eurasians and that ancestors of the Fulani probably acquired the haplotype via recent admixture events which took place in northern Africa. (...)
We screened the sequences for five variants (−13907*G, −13910*T, −13915*G, −14009*G, and −14010*C) associated with LP. The most frequent variant in our dataset, −13910*T, was found in all analyzed western Sahelian pastoralists such as the Fulani, Tuareg, and Moors in more or less similar frequencies but was not present in other groups. The second most widespread variant in our dataset, −13915*G, was found in seven populations living in the eastern part of the region. It occurred at the highest frequency among the Kababish Arabs (33.3%), while in other populations, its frequency was rather low. The third most frequent variant was −14009*G, found almost exclusively among the Beja (42.6%): in other groups, it was present at much lower frequencies. Variant −13907*G was found in seven Afar, five Beja, and one Kababish individual, and variant −14010*C was not found in our dataset at all. The LP variants of the Sahelian and neighboring populations are shown in Figure 1. (...)
Using the method of Austerlitz et al. (2003), we estimated the expansion age of −13910*T in the Fulani population at 7,534–9,686 ya, along with a slow growth rate of 1.033–1.053. (...)
Discussion
Our investigation of LP variant frequencies revealed new and interesting results related to the origins of pastoralism and subsequent gene flow between pastoralists and farmers in the Sahel/Savannah belt of Africa. We observed a clear distinction between regions west and east of Lake Chad: while variant −13910*T prevails in the western Sahel, where we found it only in pastoralists such as the Fulani, Tuareg, and Moors (it is virtually absent in sedentary farmers analyzed in our dataset), eastern Sahel appears to be much more diverse and several LP-associated variants, such as −13907*G, −13915*G, and 14010*C, are found in various populations.
The −14009*G variant has been associated with LP in eastern Africa (Jones et al., 2013; Jones et al., 2015) and frequency at which we found it in the Beja near Kassala, Sudan (42.6%) is much higher than reported by other studies. (...)
Our study shows that the age of expansion of −13910*T obtained for the Fulani pastoralists in western Sahel/Savannah belt (7,534–9,686 years ago) is much older than that obtained for the −13915*G in Arabic pastoralists in eastern Sahel/Savannah belt, when applying the same method. Priehodová et al., 2017 estimated age of −13915*G at between 1,274 and 1,782 years ago, which is consistent with the time of Arabic expansion to Africa. The estimate obtained in this study for the Fulani pastoralists conforms to estimates for introduction of pastoralism in Africa which are based on archeological evidence. (...)
This first Saharan/Sahelian influx of population with LP individuals was followed by a later migration of other pastoralists from the Arabian Peninsula throughout northern Africa and Sahara up to the Lake Chad Basin, which was led by Bedouin Arabic-speaking tribes. This migration probably introduced the −13915*G variant to the region, which originated in Arabia as an adaptation to the consumption of camel milk. (...)
It has been suggested that the Fulani acquired the −13910*T haplotype by admixing with a North African population which had some Eurasian ancestry, so that this variant in the Fulani population has the same descent as the variant found in Eurasia. Furthermore, it has been estimated that two distinct admixture events took place, one 1,828 and the other 302 years ago (Vicente et al., 2019). Our estimates of the growth and selection of −13910*T haplotype in the Fulani indicate an earlier dating than the above mentioned admixture times (Vicente et al., 2019) and are consistent rather with the estimates of Coelho et al. (2005), who proposes a period of 10,125–6,060 years ago.
The age of the second main LP Sahelian variant −13915*G, which we estimated at 1.5 ka, perfectly matches the timing of migration of Bedouin tribes from Arabia to Africa. A previous estimate of the age of −13915*G, which was up to 4 ka (Enattah et al., 2008), concerns its origin in Arabia, probably Central Arabia. From there, it expanded at the beginning of Middle Ages, therefore much later than the −13910*T variant, which was already established in the western Sahelian populations.
This discrepancy between the age estimates of −13910*T in the Fulani population can be explained by the fact that the two studies, that is, our present study and Vicente et al.'s (2019), targeted different parts of the genome (a local haplotype age of growth estimate vs. a genome-wide estimate of admixture), used different dating methods, and the samples on which they were based covered different populations. Moreover, Vicente et al. (2019) analyzed only one Fulani population (from Ziniaré, Burkina Faso), while our results are based on a larger Fulani dataset stretching geographically from Senegal to Chad. Nevertheless, we want to stress that our results do not exclude the possibility of a later admixture and introgression as suggested by Vicente et al., 2019. In fact, the TAS2R genes alleles may have also been introduced to the Fulani via recent admixture with a Eurasian population (Triska et al., 2015). In short, these results reflect the complexity of the population history of Africa. (...)
Because the −13910*T haplotype associated with LP is shared between the Fulani from Ziniaré, Burkina Faso, and Europeans (Vicente et al., 2019) and since we found an old age of expansion for this mutation within the African population of the Fulani (dataset covering much larger area of the Sahel), it is possible that the −13910*T may have originated in a food-producing population that lived in the Near East, including northeastern Africa, as suggested by Myles et al. (2005). Subsequently, though, the first Neolithic farmers who lived in the Near East/northeastern Africa were replaced by various population movements (Černý, Čížková, Poloni, Al-Meeri, & Mulligan, 2016; Vyas et al., 2016), with the result that only the originally “Arabian” –13,915*G LP variant is present in this region today (Priehodová et al., 2014). (...)
Interestingly, genetically the nearest population to the first Near Eastern farmers has been conserved in Sardinia (Chiang et al., 2018; Sikora et al., 2014), where the frequency of −13910*T is as low as elsewhere in the Mediterranean (Gerbault, 2013; Meloni et al., 2001). (...)
later migrations between Arabia, Near East, and North Africa (Černý et al., 2016; Fernandes et al., 2015) may be responsible for the fact that variant −13910*T is almost entirely absent from the Near East today (Gerbault et al., 2011)"
Last edited: