zanipolo
Banned
- Messages
- 2,071
- Reaction score
- 65
- Points
- 0
- Ethnic group
- Down Under
- Y-DNA haplogroup
- T1a2 - Z19945
- mtDNA haplogroup
- K1a4o
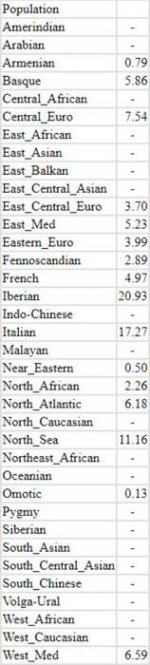
[1] "1. CLOSEST SINGLE ITEM DISTANCE%"
[1] "distance%=8.1296"
IT_Veneto,58.8
FR_Central,14
IT_Piedmont,5.8
Portugal,3
Andalusia,1.2
Sephardi_Portugal_Belmonte,1.2
Thüringen,1.2
Comunidad_Valenciana,1
FR_West,1
FR_North-East,0.8
FR_North-West,0.8
Northern_Ireland,0.8
[1] "distance%=8.1296"
IT_Veneto,58.8
FR_Central,14
IT_Piedmont,5.8
Portugal,3
Andalusia,1.2
Sephardi_Portugal_Belmonte,1.2
Thüringen,1.2
Comunidad_Valenciana,1
FR_West,1
FR_North-East,0.8
FR_North-West,0.8
Northern_Ireland,0.8