Install the app
How to install the app on iOS
Follow along with the video below to see how to install our site as a web app on your home screen.

Note: This feature currently requires accessing the site using the built-in Safari browser.
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Eurogenes Post your Eurogenes Steppe K10
- Thread starter Tomenable
- Start date
Goga
Banned
- Messages
- 2,651
- Reaction score
- 152
- Points
- 0
- Y-DNA haplogroup
- R1a*
- mtDNA haplogroup
- HV1b2
Tajiks are Iranian speaking people, they were close to Persians / Farsi folks. But they are heavily mixed with Slavic (Russian) and Turkic tribes. You can compare them to the ancient Steppe Scythians. Like the ancient 'Iraninized' Scythians of the Steppes. Scythians in the Steppes (mixture between Slavic and Turkic tribes) were 'Iranized' by East Iranian people from South East Asia (BMAC / Hindi Kush area).I noticed that Pamiri Tajiks have on average over 40% of Hindu Kush.
They also score a lot of Steppe (Yamnaya) and European Mesolithic (WHG-UHG), as well as Early Neolithic Farmer (Near Eastern).
Amazingly, they only score few % of East Asian, despite living near the border of China:
Capital of Tajikistan is Dushanbe.
Dushanbe is an Iranian/Persian/Kurdish word for 'Monday'.
"Sh (Şem.)" of the word Dushanbe (names of weekdays) has Mesopotamian roots. From
Kurdish names of weekdays
| Weekdays | Saturday | Sunday | Monday | Tuesday | Wednesday | Thursday | Friday |
|---|---|---|---|---|---|---|---|
| Rojî Hefte | Şeme | Yekşeme | Duşeme | Sêşeme | Çarşeme | Pêncşeme | Heyinî |
| Roja Heftan | Şemî | Yekşem | Duşem | Sêşem | Çarşem | Pêncşem | Înî |
| Abbreviations | Şe | Ye | Du | Sê | Ça | Pê | În |
The Kurdish name for the first day of the week Sheme (Saturday) is in fact descended from Sumerian Shabbât !!!
Greek - Sabbaton
German - Samstag
Italian - sabato
Spanish - sábado
French - Samedi
Eglish - Saturday
Dutch - zaterdag
Russian - Суббота
https://en.wikipedia.org/wiki/Kurdish_calendar
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
Goga, here is a comparison of your results with another Iranian in Steppe K10:
(BTW, your combined % of Siberian+Amerindian is similar to his East Asian):
Yes probably you are right, it might be good for Europeans but not for Iranians.
But I just wanted to check your "Afanasievo_Yamnaya" score in that calculator.
For comparing overall ancestry, I think that HarappaWorld is better for Iranians.
A comparison of your, Iran_recent's and some Iranian results in HarappaWorld:
Can you post your HarappaWorld results? I will edit this post and add them too.
(BTW, your combined % of Siberian+Amerindian is similar to his East Asian):
| Admixture: | Zoran: | Goga: |
| Near Eastern | 44,37 | 44,35 |
| Hindu Kush | 34,81 | 35,31 |
| Steppe | 12,48 | 13,35 |
| WHG-UHG | 5,22 | 2,94 |
| East Asian | 2,17 | 0,36 |
| Sub Saharan | 0,47 | 0,25 |
| Oceanian | 0,42 | 0,17 |
| Southeast Asian | 0,05 | 0,92 |
| Siberian | - | 1,23 |
| Amerindian | - | 1,11 |
Goga said:This calculator is nonsense.
Yes probably you are right, it might be good for Europeans but not for Iranians.
But I just wanted to check your "Afanasievo_Yamnaya" score in that calculator.
For comparing overall ancestry, I think that HarappaWorld is better for Iranians.
A comparison of your, Iran_recent's and some Iranian results in HarappaWorld:
| Admixture: | Zoran: | Arya: | Goga: | Iran 1400s: |
| Caucasian | 40,79 | 40,88 | ? | 45,47 |
| Baloch | 27,95 | 29,33 | ? | 26,89 |
| SW-Asian | 13,29 | 14,96 | ? | 13,13 |
| NE-Euro | 4,30 | 4,75 | ? | 7,27 |
| Mediterranean | 8,05 | 4,17 | ? | 4,87 |
| S-Indian | 2,99 | 3,71 | ? | 1,23 |
| Siberian | 0,96 | 0,22 | ? | 0,65 |
| W-African | - | - | ? | 0,20 |
| SE-Asian | - | 0,27 | ? | 0,18 |
| Beringian | - | 0,26 | ? | 0,12 |
| NE-Asian | - | 0,39 | ? | - |
| Papuan | 0,28 | 0,92 | ? | - |
| American | 0,67 | - | ? | - |
| San | 0,63 | - | ? | - |
| E-African | 0,07 | 0,14 | ? | - |
| Pygmy | - | - | ? | - |
Can you post your HarappaWorld results? I will edit this post and add them too.
Goga
Banned
- Messages
- 2,651
- Reaction score
- 152
- Points
- 0
- Y-DNA haplogroup
- R1a*
- mtDNA haplogroup
- HV1b2
Thanks, but who is this Zoran person? Is (s)he Kurdish? Zoran's auDNA is very similar to me.For comparing overall ancestry, I think that HarappaWorld is better for Iranians.
A comparison of your, Iran_recent's and some Iranian results in HarappaWorld:
...
Can you post your HarappaWorld results? I will edit this post and add them too.
My friend, first of all I'm NOT an Iranian-Persian. I'm not from Iran. I'm a 'pure' ethnic Ezdi Kurd born in Georgia (USSR). Because of my native Kurdish religion I'm not mixed with Turks or Arabs/Semites at least for the last 1000 of years. My roots are from both, North and South Kurdistan. Kurds are descendants of the Medes. 2000 years ago Persians and Medes were almost identical to each other and you can see this even nowadays. Even today there are very much similarities between Kurds (Medes) and Iranians (Persians).
But Kurds are DIFFERENT from Persians. Our languages are similar but not the same. It is like differences between Dutch, Danes and Germans..
Ethnically speaking, the closest people to Kurds are Persians, then Caucasians (Georgians, Maykop folks/Adygeans).
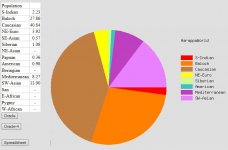
| Admixture: | Zoran: | Arya: | Goga: | Iran 1400s: |
| Caucasian | 40,79 | 40,88 | 40.84 | 45,47 |
| Baloch | 27,95 | 29,33 | 27.86 | 26,89 |
| SW-Asian | 13,29 | 14,96 | 13.9 | 13,13 |
| NE-Euro | 4,30 | 4,75 | 3.92 | 7,27 |
| Mediterranean | 8,05 | 4,17 | 8.27 | 4,87 |
| S-Indian | 2,99 | 3,71 | 2.23 | 1,23 |
| Siberian | 0,96 | 0,22 | 1.08 | 0,65 |
| W-African | - | - | - | 0,20 |
| SE-Asian | - | 0,27 | 0.57 | 0,18 |
| Beringian | - | 0,26 | - | 0,12 |
| NE-Asian | - | 0,39 | - | - |
| Papuan | 0,28 | 0,92 | 0.36 | - |
| American | 0,67 | - | 0.98 | - |
| San | 0,63 | - | - | - |
| E-African | 0,07 | 0,14 | - | - |
| Pygmy | - | - | - | - |
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
Thanks, but who is this Zoran person? Is (s)he Kurdish? Zoran's auDNA is very similar to me.
(...)
Kurds are descendants of the Medes.
Zoran is a Western Iranian and he says that his ancestors were Medes and Parthians.
Kurds and Iranians are very similar.
Also Armenians are close.Ethnically speaking, the closest people to Kurds are Persians, then Caucasians (Georgians, Maykop folks/Adygeans).
Goga
Banned
- Messages
- 2,651
- Reaction score
- 152
- Points
- 0
- Y-DNA haplogroup
- R1a*
- mtDNA haplogroup
- HV1b2
Close, but not that close. Armenians are quite different. Armenians are more shifted toward the Levant, Med. Sea. Armenians match better with (and closer to) Druze and Cypriots, than with Kurds, Iranians and Adygeians.Also Armenians are close.
+ Armenian are closer to Georgians than to Iranians, Kurds and Adygeians
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
I compared my results with RISE150 - who lived in what is now Poland, but ~3900 years ago:
I used calculator puntDNAL K15 for this comparison:
1) My result:
Single Population Sharing:
# Population (source) Distance
1 Polish 2.06
2 Swedish 4.03
3 Norwegian 6.35
4 North_German 6.79
5 Belarusian 7.58
6 Slovenian 7.94
2) RISE 150:
Single Population Sharing:
# Population (source) Distance
1 Swedish 5.58
2 North_German 5.95
3 Polish 6.07
4 Slovenian 6.29
5 Norwegian 6.58
6 Hungarian 7.41
She has the same populations as top results in "population sharing" - only in different order.
This cannot be coincidence. RISE150 was a sample of Unetice culture:
http://www.ancestraljourneys.org/copperbronzeagedna.shtml
RISE150 is available on GEDmatch (kit number F999948):
http://www.y-str.org/p/ancient-dna.html
I used calculator puntDNAL K15 for this comparison:
1) My result:
Single Population Sharing:
# Population (source) Distance
1 Polish 2.06
2 Swedish 4.03
3 Norwegian 6.35
4 North_German 6.79
5 Belarusian 7.58
6 Slovenian 7.94
2) RISE 150:
Single Population Sharing:
# Population (source) Distance
1 Swedish 5.58
2 North_German 5.95
3 Polish 6.07
4 Slovenian 6.29
5 Norwegian 6.58
6 Hungarian 7.41
She has the same populations as top results in "population sharing" - only in different order.
This cannot be coincidence. RISE150 was a sample of Unetice culture:
http://www.ancestraljourneys.org/copperbronzeagedna.shtml
RISE150 is available on GEDmatch (kit number F999948):
http://www.y-str.org/p/ancient-dna.html
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
Please note, that:
These GEDmatch calculators (such as puntDNAL K15 used above) are good in detecting even very old admixtures, from several thousand years ago. On the other hand, ancestry reports from DNALand, 23andMe or FTDNA are only accurate in showing ancestry no more than up to one thousand years ago. They are good for recent history.
This is why I was very suprised when puntDNAL K15 Oracle showed that I have similar admixture proportions to Swedes, even though according to FTDNA I do not have any Scandinavian admixture.
I understood what is going on only after comparing my results to those of RISE150.
These admixtures have been present here already in the Bronze Age!
====================
My FTDNA report - no any Scandinavian admixture, only Eastern + Central Euro:
(but as I wrote, this report covers accurately only the last 1000 years, at best)

These GEDmatch calculators (such as puntDNAL K15 used above) are good in detecting even very old admixtures, from several thousand years ago. On the other hand, ancestry reports from DNALand, 23andMe or FTDNA are only accurate in showing ancestry no more than up to one thousand years ago. They are good for recent history.
This is why I was very suprised when puntDNAL K15 Oracle showed that I have similar admixture proportions to Swedes, even though according to FTDNA I do not have any Scandinavian admixture.
I understood what is going on only after comparing my results to those of RISE150.
These admixtures have been present here already in the Bronze Age!
====================
My FTDNA report - no any Scandinavian admixture, only Eastern + Central Euro:
(but as I wrote, this report covers accurately only the last 1000 years, at best)

LeBrok
Elite member
- Messages
- 10,261
- Reaction score
- 1,617
- Points
- 0
- Location
- Calgary
- Ethnic group
- Citizen of the world
- Y-DNA haplogroup
- R1b Z2109
- mtDNA haplogroup
- H1c
I compared my results with RISE150 - who lived in what is now Poland, but ~3900 years ago:
I used calculator puntDNAL K15 for this comparison:
1) My result:
Single Population Sharing:
# Population (source) Distance
1 Polish 2.06
2 Swedish 4.03
3 Norwegian 6.35
4 North_German 6.79
5 Belarusian 7.58
6 Slovenian 7.94
2) RISE 150:
Single Population Sharing:
# Population (source) Distance
1 Swedish 5.58
2 North_German 5.95
3 Polish 6.07
4 Slovenian 6.29
5 Norwegian 6.58
6 Hungarian 7.41
She has the same populations as top results in "population sharing" - only in different order.
This cannot be coincidence. RISE150 was a sample of Unetice culture:
http://www.ancestraljourneys.org/copperbronzeagedna.shtml
RISE150 is available on GEDmatch (kit number F999948):
http://www.y-str.org/p/ancient-dna.html
They are not too much off, and point to genetic stability since Bronze age in this area. However, do we see a little shift to the East, to Slavic nations?
Do we have any samples from 500BC to 500AD, to see how it looked before Slavic expansion? To have a look at these "Veneti" or Germanic expansion in the area.
| 1 | Polish | 1.79 |
| 2 | Swedish | 4.49 |
| 3 | Belarusian | 6.58 |
| 4 | Norwegian | 6.72 |
| 5 | North_German | 7.69 |
| 6 | Russian | 8.03 |
Twilight
Regular Member
- Messages
- 956
- Reaction score
- 91
- Points
- 28
- Location
- Clinton, Washington
- Ethnic group
- 15/32 British, 5/32 German, 9/64 Irish, 1/8 Scots Gaelic, 5/64 French, 1/32 Welsh
- Y-DNA haplogroup
- R1b-U152-Z56-BY3957
- mtDNA haplogroup
- J1c7a
Sorry! Wrong link.
SteppeK10: https://drive.google.com/file/d/0B-XBmvmgdkfVM2RRRHlRSkcwV0k/view
I used R software for Windows. See the "readme": https://drive.google.com/file/d/0B7AJcY18g2GaZGU4OWQ5OWItMzY2NC00NzI1LWIzNWMtMzUxYWI4NjRmMTlk/view
A summary based on the readme file:
1 - Download, install and run R software.
2 - "Once R is running, it will give you a command prompt where you can enter commands. First, you must change the directory to your working directory.
You can do this from the File -> Change dir menu in Windows"
3 - At the R prompt, enter:
source('standardize.r')
"This loads a small program that will convert your data from the
company-specific format to a common format in the next step."
4 - At the R prompt, enter:
a. If you have 23andMe data (either v2 or v3 chip):
standardize('yourgenome.txt', company='23andMe')
b. if you have Family Finder data (Illumina chip only):
standardize('yourgenome.csv', company='ftdna')
5 - At the R prompt, enter:
system('DIYDodecadWin steppe.par')
Im afraid my IPad can't download using the R software. Do you knowing there is a way for my IPad to download Steepes10?
LeBrok
Elite member
- Messages
- 10,261
- Reaction score
- 1,617
- Points
- 0
- Location
- Calgary
- Ethnic group
- Citizen of the world
- Y-DNA haplogroup
- R1b Z2109
- mtDNA haplogroup
- H1c
It is not going well for me either, I've downloaded all but can't quickly figure out all the necessary steps. Foreign concept for me to run programs in a program and have with weird names to the right folders. I don't want to spend hours with this to experiment endlessly what works. I need baby steps manual, if someone cares to right them down.Im afraid my IPad can't download using the R software. Do you knowing there is a way for my IPad to download Steepes10?
LeBrok
Elite member
- Messages
- 10,261
- Reaction score
- 1,617
- Points
- 0
- Location
- Calgary
- Ethnic group
- Citizen of the world
- Y-DNA haplogroup
- R1b Z2109
- mtDNA haplogroup
- H1c
What is very interesting is the fact that Rise 150 is almost at equal distance to modern populations of Germanics and Slavs of the Area. All from 5.58 to 6.58, that's a very small margin to fit all in. This must be true Corded Ware heritage for both groups.I compared my results with RISE150 - who lived in what is now Poland, but ~3900 years ago:
2) RISE 150:
Single Population Sharing:
# Population (source) Distance
1 Swedish 5.58
2 North_German 5.95
3 Polish 6.07
4 Slovenian 6.29
5 Norwegian 6.58
6 Hungarian 7.41
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
Now I tried 1-to-1 comparison:
Comparing Kit T269964 (*Tomenable) and F999948 (RISE150,Poland,3.4ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 4.7 cM
Total of segments > 1 cM = 1,142.2 cM
701 matching segments
307164 SNPs used for this comparison.
Comparison took 0.03335 seconds.
===================================
Comparing Kit T269964 (*Tomenable) and F999933 (BR2, Hungary, 3.2ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 6.8 cM
Total of segments > 1 cM = 1,488.4 cM
921 matching segments
657082 SNPs used for this comparison.
Comparison took 0.04641 seconds.
=============
=============
I have 701 matching segments with RISE150 and a bit more (921) with Hungarian BR2.
However, Hungarian BR2 is not similar to me and to RISE150 in "Single Population Sharing".
In puntDNAL K15:
3) BR2 (Kyjatice culture):
Single Population Sharing:
# Population (source) Distance
1 Utahn_White 4.41
2 South_German 4.52
3 French 5.02
4 English 6
5 Orcadian 6.91
6 Scottish 7.3
7 Irish 7.61
BR2 has totally different populations (no Poles, no Swedes, no Slovenians, no Norwegians, etc.).
I'm not sure how to interpret this (why more of matching segments, if less matching populations?).
Comparing Kit T269964 (*Tomenable) and F999948 (RISE150,Poland,3.4ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 4.7 cM
Total of segments > 1 cM = 1,142.2 cM
701 matching segments
307164 SNPs used for this comparison.
Comparison took 0.03335 seconds.
===================================
Comparing Kit T269964 (*Tomenable) and F999933 (BR2, Hungary, 3.2ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 6.8 cM
Total of segments > 1 cM = 1,488.4 cM
921 matching segments
657082 SNPs used for this comparison.
Comparison took 0.04641 seconds.
=============
=============
I have 701 matching segments with RISE150 and a bit more (921) with Hungarian BR2.
However, Hungarian BR2 is not similar to me and to RISE150 in "Single Population Sharing".
In puntDNAL K15:
3) BR2 (Kyjatice culture):
Single Population Sharing:
# Population (source) Distance
1 Utahn_White 4.41
2 South_German 4.52
3 French 5.02
4 English 6
5 Orcadian 6.91
6 Scottish 7.3
7 Irish 7.61
BR2 has totally different populations (no Poles, no Swedes, no Slovenians, no Norwegians, etc.).
I'm not sure how to interpret this (why more of matching segments, if less matching populations?).
LeBrok
Elite member
- Messages
- 10,261
- Reaction score
- 1,617
- Points
- 0
- Location
- Calgary
- Ethnic group
- Citizen of the world
- Y-DNA haplogroup
- R1b Z2109
- mtDNA haplogroup
- H1c
Because the other populations are even closer to him than you. For example French can have: Total of segments > 1 cM = 1,600 cM and 1002 matching segmentsNow I tried 1-to-1 comparison:
Comparing Kit T269964 (*Tomenable) and F999948 (RISE150,Poland,3.4ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 4.7 cM
Total of segments > 1 cM = 1,142.2 cM
701 matching segments
307164 SNPs used for this comparison.
Comparison took 0.03335 seconds.
===================================
Comparing Kit T269964 (*Tomenable) and F999933 (BR2, Hungary, 3.2ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 6.8 cM
Total of segments > 1 cM = 1,488.4 cM
921 matching segments
657082 SNPs used for this comparison.
Comparison took 0.04641 seconds.
=============
=============
I have 701 matching segments with RISE150 and a bit more (921) with Hungarian BR2.
However, Hungarian BR2 is not similar to me and to RISE150 in "Single Population Sharing".
In puntDNAL K15:
3) BR2 (Kyjatice culture):
Single Population Sharing:
# Population (source) Distance
1 Utahn_White 4.41
2 South_German 4.52
3 French 5.02
4 English 6
5 Orcadian 6.91
6 Scottish 7.3
7 Irish 7.61
BR2 has totally different populations (no Poles, no Swedes, no Slovenians, no Norwegians, etc.).
I'm not sure how to interpret this (why more of matching segments, if less matching populations?).
I wonder if BR2 is the source of Celtic type IEs?
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
I carried out also a 1-to-1 comparison with Hinxton-4 (Iron Age Briton) - just to make sure that I'm not as closely related to Ancient Britons as to RISE150 (which would imply that there is nothing special about this similarity with RISE150):
Comparing Kit T269964 (*Tomenable) and F999925 (Hinxton-4, UK, 2ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 2.7 cM
Total of segments > 1 cM = 162.9 cM
125 matching segments
575225 SNPs used for this comparison.
Comparison took 0.02972 seconds.
========================
Only 125 segments, much fewer than with RISE150 and BR2.
Comparing Kit T269964 (*Tomenable) and F999925 (Hinxton-4, UK, 2ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 2.7 cM
Total of segments > 1 cM = 162.9 cM
125 matching segments
575225 SNPs used for this comparison.
Comparison took 0.02972 seconds.
========================
Only 125 segments, much fewer than with RISE150 and BR2.
LeBrok
Elite member
- Messages
- 10,261
- Reaction score
- 1,617
- Points
- 0
- Location
- Calgary
- Ethnic group
- Citizen of the world
- Y-DNA haplogroup
- R1b Z2109
- mtDNA haplogroup
- H1c
That's a bit weird perhaps, but keep in mind that matches will be fewer if we are not dealing with the whole genome. Many ancient genomes are only partial.I carried out also a 1-to-1 comparison with Hinxton-4 (Iron Age Briton) - just to make sure that I'm not as closely related to Ancient Britons as to RISE150 (which would imply that there is nothing special about this similarity with RISE150):
Comparing Kit T269964 (*Tomenable) and F999925 (Hinxton-4, UK, 2ky):
Minimum threshold size to be included in total = 25 SNPs
Mismatch-bunching Limit = 25 SNPs
Minimum segment cM to be included in total = 1.0 cM
Largest segment = 2.7 cM
Total of segments > 1 cM = 162.9 cM
125 matching segments
575225 SNPs used for this comparison.
Comparison took 0.02972 seconds.
========================
Only 125 segments, much fewer than with RISE150 and BR2.
Fluffy
Regular Member
- Messages
- 186
- Reaction score
- 28
- Points
- 0
- Ethnic group
- Dutch / French
- Y-DNA haplogroup
- G2a l497 s10458
- mtDNA haplogroup
- H1n
It is not going well for me either, I've downloaded all but can't quickly figure out all the necessary steps. Foreign concept for me to run programs in a program and have with weird names to the right folders. I don't want to spend hours with this to experiment endlessly what works. I need baby steps manual, if someone cares to right them down.
LOL same here man. I can't figure this out unless I am willing to spend hours to study it.
Tomenable
Elite member
- Messages
- 5,419
- Reaction score
- 1,337
- Points
- 113
- Location
- Poland
- Ethnic group
- Polish
- Y-DNA haplogroup
- R1b-L617
- mtDNA haplogroup
- W6a
Some more ancients in puntDNAL K15 calculator:
Iron Age Celtic Briton was most similar to modern Irish, Anglo-Saxon to modern Swedes:
Hinxton-4 (Celtic Briton, 2000 ybp):
Kit F999925. Single Population Sharing:
# Population (source) Distance
1 Irish 2.38
2 North_German 2.95
3 Orcadian 3.03
4 English 3.05
5 Scottish 3.18
6 Norwegian 4.4
7 Slovenian 4.44
8 Austrian 4.91
Hinxton-2 (Anglo-Saxon, 1300 ybp):
Kit F999921. Single Population Sharing:
# Population (source) Distance
1 Swedish 4.3
2 Norwegian 4.41
3 Scottish 5.73
4 North_German 5.87
5 Polish 6.44
6 Orcadian 6.82
7 English 7.24
8 Irish 7.93
==========================
It is quite surprising, that North Germans are similar both to Celtic Briton & to Anglo-Saxon.
Even more surprising, that Germans are more similar to Celtic Briton than to Saxon.
Perhaps she (Hinxton-2) was not even a Saxon, but either an Angle or a Jute.
==========================
Modern English are closer to Celtic Briton (distance 3.05) than to unmixed Anglo-Jute-Saxon (distance 7.24). This is in agreement with recent studies (from 2015-2016) which showed that the English have mostly Celtic / Briton ancestry.
"Pure Anglo-Saxon-Jutes" were Scandinavian-like rather than English-like or German-like.
This also shows why it is hard to distinguish Anglo-Saxon-Jute and Danish-Swedish Viking ancestry.
Only Norwegian Viking ancestry can be distinguished, because Norwegians are a bit different.
Iron Age Celtic Briton was most similar to modern Irish, Anglo-Saxon to modern Swedes:
Hinxton-4 (Celtic Briton, 2000 ybp):
Kit F999925. Single Population Sharing:
# Population (source) Distance
1 Irish 2.38
2 North_German 2.95
3 Orcadian 3.03
4 English 3.05
5 Scottish 3.18
6 Norwegian 4.4
7 Slovenian 4.44
8 Austrian 4.91
Hinxton-2 (Anglo-Saxon, 1300 ybp):
Kit F999921. Single Population Sharing:
# Population (source) Distance
1 Swedish 4.3
2 Norwegian 4.41
3 Scottish 5.73
4 North_German 5.87
5 Polish 6.44
6 Orcadian 6.82
7 English 7.24
8 Irish 7.93
==========================
It is quite surprising, that North Germans are similar both to Celtic Briton & to Anglo-Saxon.
Even more surprising, that Germans are more similar to Celtic Briton than to Saxon.
Perhaps she (Hinxton-2) was not even a Saxon, but either an Angle or a Jute.
==========================
Modern English are closer to Celtic Briton (distance 3.05) than to unmixed Anglo-Jute-Saxon (distance 7.24). This is in agreement with recent studies (from 2015-2016) which showed that the English have mostly Celtic / Briton ancestry.
"Pure Anglo-Saxon-Jutes" were Scandinavian-like rather than English-like or German-like.
This also shows why it is hard to distinguish Anglo-Saxon-Jute and Danish-Swedish Viking ancestry.
Only Norwegian Viking ancestry can be distinguished, because Norwegians are a bit different.
Twilight
Regular Member
- Messages
- 956
- Reaction score
- 91
- Points
- 28
- Location
- Clinton, Washington
- Ethnic group
- 15/32 British, 5/32 German, 9/64 Irish, 1/8 Scots Gaelic, 5/64 French, 1/32 Welsh
- Y-DNA haplogroup
- R1b-U152-Z56-BY3957
- mtDNA haplogroup
- J1c7a
It is not going well for me either, I've downloaded all but can't quickly figure out all the necessary steps. Foreign concept for me to run programs in a program and have with weird names to the right folders. I don't want to spend hours with this to experiment endlessly what works. I need baby steps manual, if someone cares to right them down.
LOL same here man. I can't figure this out unless I am willing to spend hours to study it.
Im glad I'm not the only one, hopefully our personal results will come to us soon.
Best of luck to Davidski, it appears that near Eastern genome among Bedouins in 60% and 2% WHG so SteepesK10 sounds promising.
LeBrok
Elite member
- Messages
- 10,261
- Reaction score
- 1,617
- Points
- 0
- Location
- Calgary
- Ethnic group
- Citizen of the world
- Y-DNA haplogroup
- R1b Z2109
- mtDNA haplogroup
- H1c
As I said before, perhaps it depends how much ancient DNA was recovered? Most of them are partial.I share 701 matching segments with RISE150 and 921 with BR2.
Numbers such as 701 - 921 matching segments seem to be a lot.
I made a similar comparison with ancient and modern Iranians:
Kit T637158 (Medieval Iranian) shares 758 segments with a modern Iranian.
Kit M124870 (Copper Age Iranian) shares 597 segments with a modern Iranian.
Finally, T637158 and M124870 share 380 matching segments with each other.
Criteria of comparison were the same in each case:
This thread has been viewed 49508 times.
