paul333
Banned
- Messages
- 221
- Reaction score
- 32
- Points
- 0
- Y-DNA haplogroup
- H2a1 M9313
- mtDNA haplogroup
- H1c3
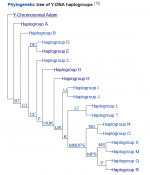
I have been looking into my raw Data results,from living Dna, and have found each SNP, I am positive for on My 'Y Data' , identifies a main Haplogroup. Is this identifying an ancestor from this group. I have 199 positive SNPs confirmed in my Y- raw data, is this or can this identifying SNP marker specificly relate to an individual Y ancestor. ?
Last edited: