Salento
Regular Member
- Messages
- 5,635
- Reaction score
- 2,838
- Points
- 113
- Ethnic group
- Italian
- Y-DNA haplogroup
- T1a2 - SK1480
- mtDNA haplogroup
- H12a
:cool-v: @Duarte
your mtDNA seems to trace the entire migration into America.
You mentioned 45.000 year old Ust'-Ishim Man, one of the early modern humans to inhabit Western Siberia, y K-M526, mtDNA R
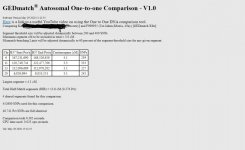
Ust'-Ishim GedMatch # F999935
one to one comparison:
S vs Ust'-Ishim
Larg. Segm = 3.5 cM - (HIR) = 10.2 cM

your mtDNA seems to trace the entire migration into America.
You mentioned 45.000 year old Ust'-Ishim Man, one of the early modern humans to inhabit Western Siberia, y K-M526, mtDNA R
Ust'-Ishim GedMatch # F999935
one to one comparison:
S vs Ust'-Ishim
Larg. Segm = 3.5 cM - (HIR) = 10.2 cM