kingjohn
Regular Member
- Messages
- 2,229
- Reaction score
- 1,198
- Points
- 113
It came from the steppe present in both Italics and Etruscans.
Nothing at all to do with Slavs.
ok ,
thats what i thought
the most logical explanation
Follow along with the video below to see how to install our site as a web app on your home screen.

Note: This feature currently requires accessing the site using the built-in Safari browser.
It came from the steppe present in both Italics and Etruscans.
Nothing at all to do with Slavs.
Thank you for getting involved, Pax! Really useful information.Baltic is already present in Etruscans and Latins. Were the Etruscans and Latins also descended from Slavic migrations that are later than them? It seems clear to me that the Baltic = Slavic equation is wrong.
R1015_Lazio_Rome_Iron_Age_Villanovan,34.16,9.13,33.08,0.00,20.98,1.79,0.00,0.27,0.00,0.59,0.00,0.00,0.00
R473_Civitavecchia_Etruscan._Iron_Age,33.38,12.03,31.63,0.41,18.38,3.34,0.34,0.42,0.06,0.00,0.00,0.00,0.00
R474b_Civitavecchia_Etruscan._Iron_Age,35.67,12.22,25.57,5.16,17.48,1.51,0.00,0.00,1.10,0.59,0.00,0.71,0.00
R1016_Lazio_Rome_Iron_Age_Latini,35.91,7.69,33.00,1.06,20.82,0.00,0.00,0.00,0.00,1.52,0.00,0.00,0.00
R1021_Lazio_Frosinone_Iron_Age_Latini,37.21,9.04,30.58,2.18,18.82,1.44,0.00,0.04,0.00,0.00,0.70,0.00,0.00
R851_Lazio_Rome_Iron_Age_Latini,34.22,14.60,34.26,1.01,15.44,0.30,0.00,0.00,0.18,0.00,0.00,0.00,0.00
Someone is there with possible haplogroups associated with Slavs but it is a small minority, nothing that can justify a mass replacement.
| Target: CZE_Early_Slav Distance: 2.7478% / 0.02747845 | ADC: 0.25x RC | |
|---|---|
| 46.0 | HUN_MA_Szolad |
| 40.2 | HUN_Avar_Szolad |
| 13.8 | Baltic_EST_BA |
| Target: Belarusian Distance: 1.3935% / 0.01393459 | ADC: 0.25x RC | |
|---|---|
| 63.2 | HUN_Avar_Szolad |
| 24.6 | Baltic_EST_BA |
| 12.2 | CZE_Early_Slav |
| Target: Serbian Distance: 3.7958% / 0.03795817 | ADC: 0.25x RC | |
|---|---|
| 66.2 | HUN_MA_Szolad |
| 33.8 | CZE_Early_Slav |
| Target: Italian_Lombardy Distance: 6.1244% / 0.06124430 | ADC: 0.25x RC | |
|---|---|
| 100.0 | HUN_MA_Szolad |
| Target: Italian_Tuscany Distance: 6.9928% / 0.06992756 | ADC: 0.25x RC | |
|---|---|
| 100.0 | HUN_MA_Szolad |
| Target: Greek_Central_Macedonia Distance: 7.0482% / 0.07048239 | ADC: 0.25x RC | |
|---|---|
| 100.0 | HUN_MA_Szolad |
| Target: Albanian Distance: 7.1215% / 0.07121452 | ADC: 0.25x RC | |
|---|---|
| 100.0 | HUN_MA_Szolad |
| Target: CZE_Early_Slav Distance: 2.7478% / 0.02747845 | ADC: 0.25x RC | |
|---|---|
| 46.0 | HUN_MA_Szolad |
| 40.2 | HUN_Avar_Szolad |
| 13.8 | Baltic_EST_BA |
| Target: Belarusian Distance: 1.3935% / 0.01393459 | ADC: 0.25x RC | |
|---|---|
| 63.2 | HUN_Avar_Szolad |
| 24.6 | Baltic_EST_BA |
| 12.2 | CZE_Early_Slav |
| Target: Belarusian Distance: 1.3935% / 0.01393459 | ADC: 0.25x RC | |
|---|---|
| 63.2 | HUN_Avar_Szolad |
| 24.6 | Baltic_EST_BA |
| 12.2 | CZE_Early_Slav |
It came from the steppe present in both Italics and Etruscans.
Nothing at all to do with Slavs.
so is it a native elment to north east italy ?
or it arrived with germanic tribes ?
or others ?
@kingjohn
check the admixture of the lombards
you will find they are more associated with east-germanic then with west-germanic
thats why i think they probably also increased the baltic in the north of italy
afcorse it was minimal compare to the steppe movement
i understand from angela that there( LOMBARDS) y haplogroup was r-u106
but here i speak about autosomal influence not y haplogroup influence
p.s
by the way in northwest italy the baltic component is also between 10-16%

The maps on this page represents the distribution of Human Y-chromosomal DNA (Y-DNA) haplogroups. A Y-DNA haplogroup is a group of men sharing the same series of mutations on their Y chromosome, which they inherited from a long line of common paternal ancestors. A few new mutations, known as SNP's, happen every generation. This allows to retrace the genealogical tree of humanity with great accuracy and see patterns in the distribution of shared historical lineages. Most major haplogroups are many thousands of years old, typically going back to the Bronze Age, Neolithic, Mesolithic or even Paleolithic. The deeper the subclade the more recent the shared ancestor. Classifying SNP's into a genealogical order is known as phylogenentics. Phylogenetic trees of European haplogroups are available here. The following maps were compiled with all the latest available Y-DNA data for each country. The maps are frequently updated when new data becomes available.
Haplogroup I (Y-DNA) Distribution map of haplogroup I in Europe 
Haplogroup I1 (Y-DNA) Distribution map of haplogroup I1 in Europe 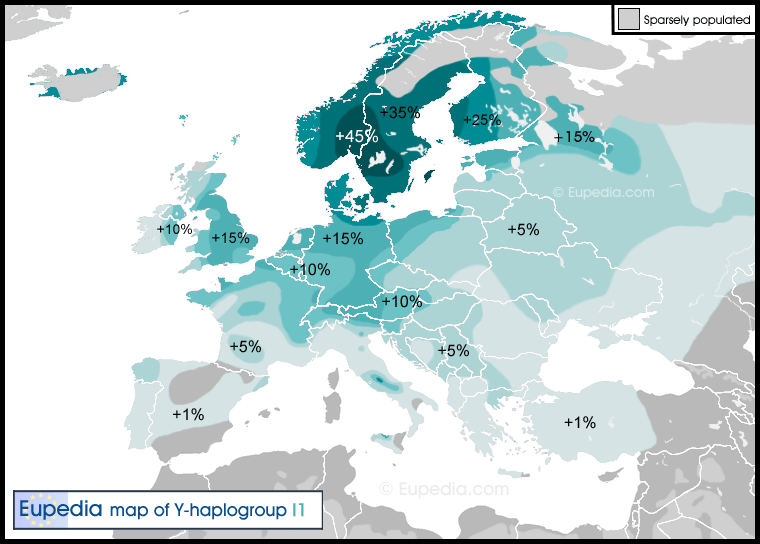
Haplogroup I2a1 (Y-DNA) Distribution map of haplogroup I2a1 in Europe 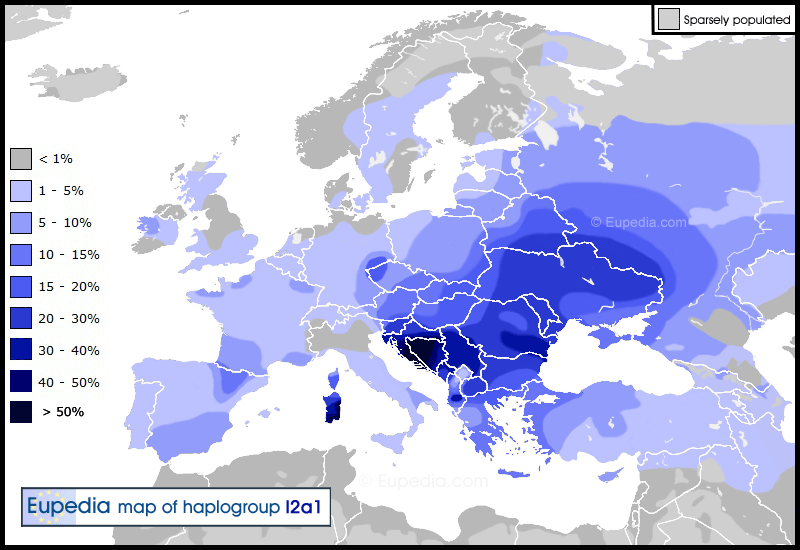
Haplogroup I2a2 (Y-DNA) Distribution map of haplogroup I2a2 in Europe 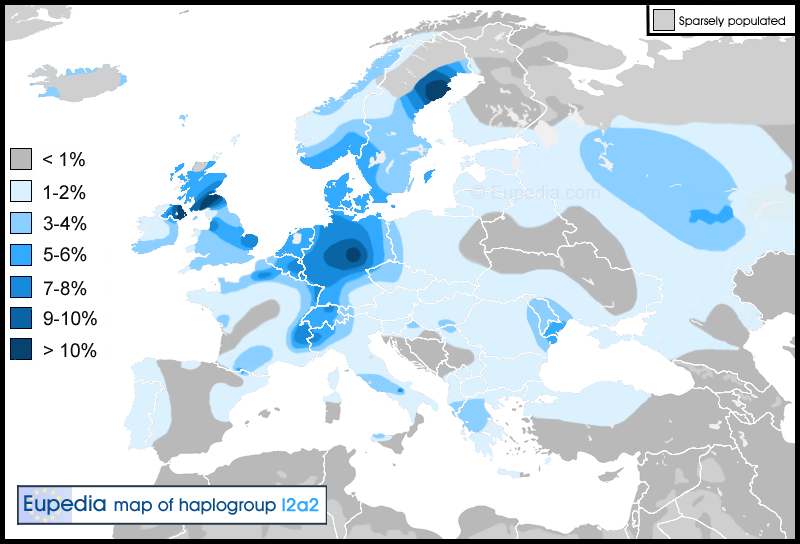
Haplogroup R1 (Y-DNA) Distribution map of haplogroup R1 in Europe 
Haplogroup R1a (Y-DNA) Distribution map of haplogroup R1a in Europe, the Middle East & North Africa 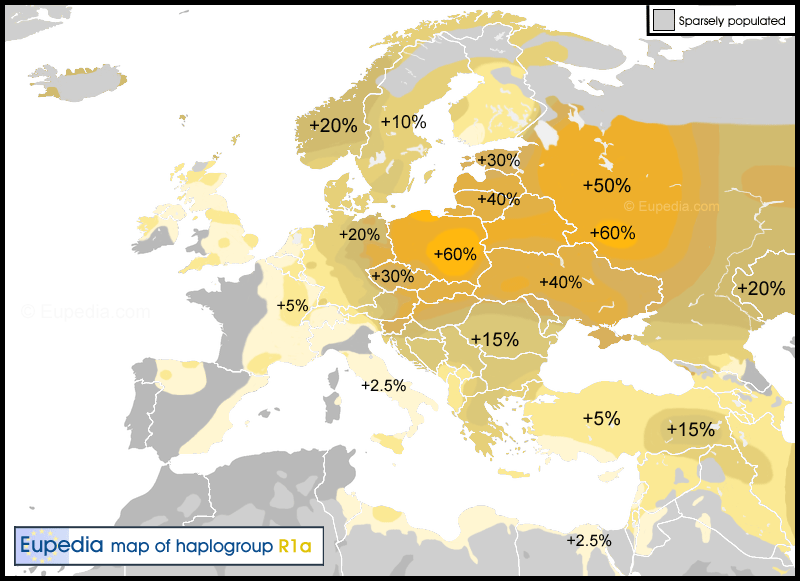
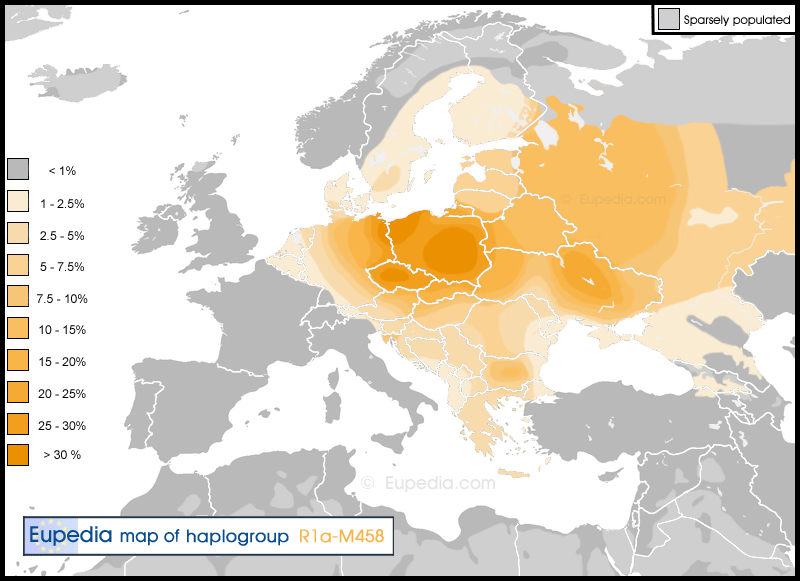

 Haplogroup R1b (Y-DNA) Distribution map of haplogroup R1b in Europe, the Middle East & North Africa 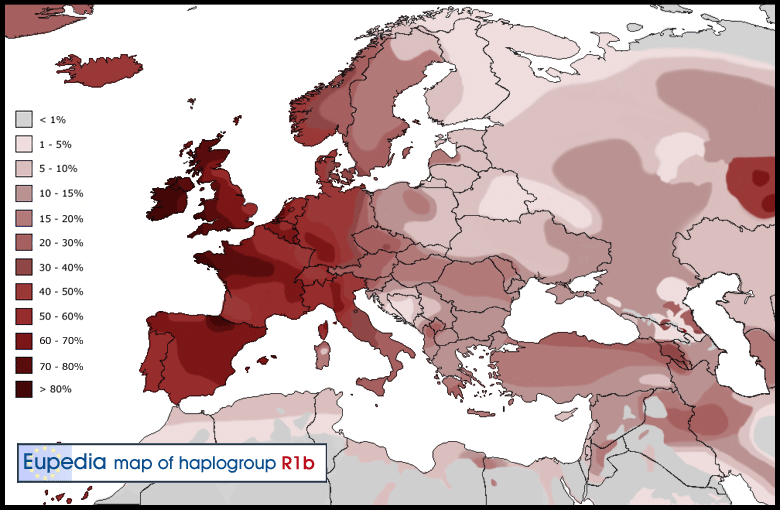 Distribution map of haplogroup R1b-S21 (U106) in Europe 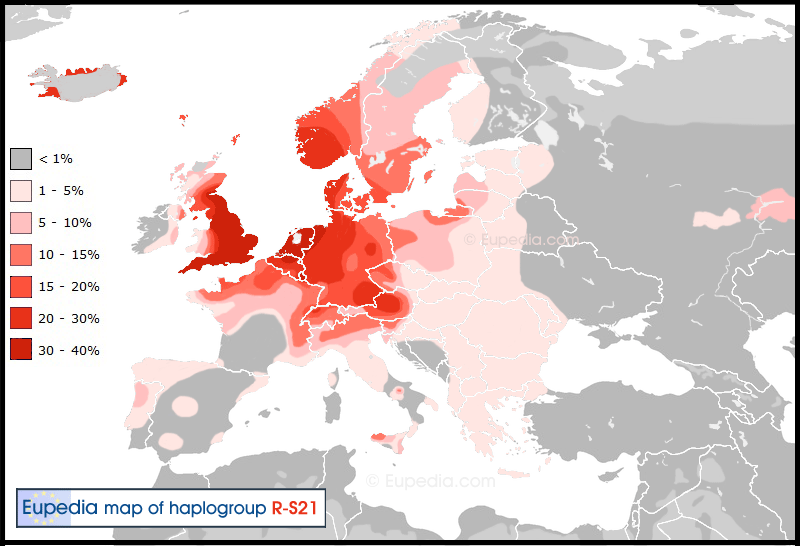
|
@kingjohn
check the admixture of the lombards
you will find they are more associated with east-germanic then with west-germanic
slavic is only linked directly with the border area of Ukraine and Belarus .................all other slavic is linked with lingustic association and not genetic association

| Target: French_Seine-Maritime Distance: 4.5734% / 0.04573431 | ADC: 0.25x RC | |
|---|---|
| 73.8 | CZE_Early_Slav |
| 26.2 | BGR_IA |
| Target: French_Provence Distance: 3.7593% / 0.03759307 | ADC: 0.25x RC | |
|---|---|
| 50.0 | BGR_IA |
| 50.0 | CZE_Early_Slav |
| Target: French_Pas-de-Calais Distance: 4.9056% / 0.04905648 | ADC: 0.25x RC | |
|---|---|
| 72.2 | CZE_Early_Slav |
| 27.8 | BGR_IA |
| Target: French_Paris Distance: 4.9416% / 0.04941613 | ADC: 0.25x RC | |
|---|---|
| 65.8 | CZE_Early_Slav |
| 34.2 | BGR_IA |
| Target: French_Occitanie Distance: 4.9053% / 0.04905323 | ADC: 0.25x RC | |
|---|---|
| 59.2 | CZE_Early_Slav |
| 40.8 | BGR_IA |
| Target: French_Nord Distance: 4.3426% / 0.04342611 | ADC: 0.25x RC | |
|---|---|
| 69.4 | CZE_Early_Slav |
| 30.6 | BGR_IA |




No matter how you look at it... the Danubian lime paper methodology is trash, what I said by taking one look at it. And I believe Pax also had some similar feelings without further specifying what at the time.
https://imgur.com/a/jbYPD4H
Interesting:cool-v:

No matter how you look at it... the Danubian lime paper methodology is trash, what I said by taking one look at it. And I believe Pax also had some similar feelings without further specifying what at the time.
https://imgur.com/a/jbYPD4H
Interesting:cool-v:
Can you try to model aschenazi jews( moldovan jews) and cretans
With this combination of
iron age bulgaria+ early slav CZE ?

Basically you can mix and match any nationality as the source combination.Sadly couldn't find the coordinates for Moldovan Jew.
But for your enjoyment :cool-v:
https://imgur.com/a/8SGIygh
You could probably guess my train of thought (algorithm) slide to slide.
This is after cleaning up the model, removing the marginally improving populations and also removing any modern populations.
I can explain the alorithm I used to come to this final table but doubt anyone much cares.

Sadly couldn't find the coordinates for Moldovan Jew.
But for your enjoyment :cool-v:
https://imgur.com/a/8SGIygh
You could probably guess my train of thought (algorithm) slide to slide.
This is after cleaning up the model, removing the marginally improving populations and also removing any modern populations.
I can explain the alorithm I used to come to this final table but doubt anyone much cares.

This thread has been viewed 20305 times.
