Syd
Regular Member
- Messages
- 46
- Reaction score
- 31
- Points
- 18
- Location
- Italy
- Ethnic group
- Latin
- Y-DNA haplogroup
- R-DF13*
- mtDNA haplogroup
- H
Abstract
The Middle to Upper Palaeolithic transition in Europe is associated with the regional disappearance of Neanderthals and the spread of Homo sapiens. Late Neanderthals persisted in western Europe several millennia after the occurrence of H. sapiens in eastern Europe1. Local hybridization between the two groups occurred2, but not on all occasions3. Archaeological evidence also indicates the presence of several technocomplexes during this transition, complicating our understanding and the association of behavioural adaptations with specific hominin groups4. One such technocomplex for which the makers are unknown is the Lincombian–Ranisian–Jerzmanowician (LRJ), which has been described in northwestern and central Europe5,6,7,8. Here we present the morphological and proteomic taxonomic identification, mitochondrial DNA analysis and direct radiocarbon dating of human remains directly associated with an LRJ assemblage at the site Ilsenhöhle in Ranis (Germany). These human remains are among the earliest directly dated Upper Palaeolithic H. sapiens remains in Eurasia. We show that early H. sapiens associated with the LRJ were present in central and northwestern Europe long before the extinction of late Neanderthals in southwestern Europe. Our results strengthen the notion of a patchwork of distinct human populations and technocomplexes present in Europe during this transitional period.
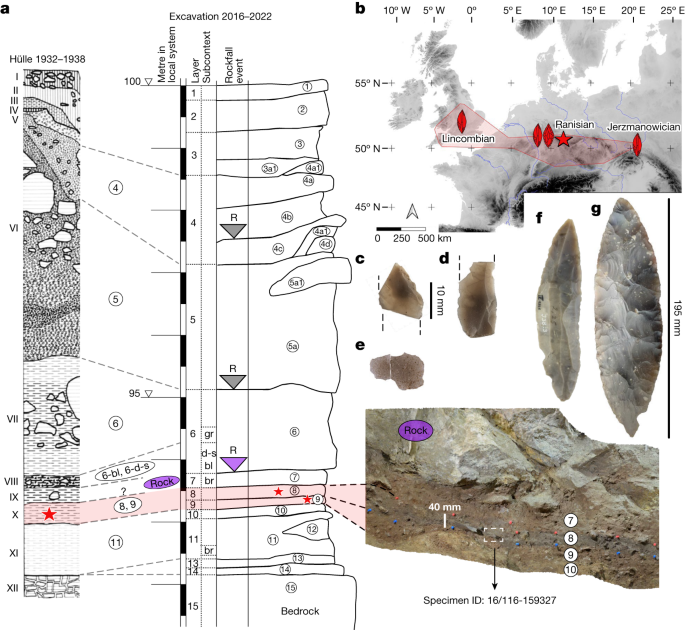
We tested 11 of the hominin remains for the preservation of ancient mitochondrial DNA (mtDNA). Between 4,413 and 175,688 unique reads mapping to the human mtDNA reference genome were recovered per skeletal fragment. These mtDNA reads had elevated frequencies of cytosine (C)-to-thymine (T) substitutions (32.6% to 49.6% on the 5′ end and 19.0% to 47.9% on the 3′ end, respectively; Supplementary Figs. 14–24), which are indicative of ancient DNA. Positions shown to be informative for differentiating between H. sapiens, Neanderthal and Denisovan mtDNA genomes enabled us to identify each of the 11 skeletal fragments as belonging to ancient H. sapiens (Supplementary Table 18). Libraries from ten of the eleven skeletal fragments contained sufficient data for reconstructing near-complete mtDNA genomes. Five of these mtDNA genomes showed no pairwise differences among them for the positions covered, suggesting that they stemmed from either the same individual or maternally related individuals (Supplementary Fig. 25 and Supplementary Table 19). Four of these skeletal fragments come from the 1932–1938 collection and one from the 2016–2022 excavation (16/116-159327; Fig. 1), providing additional support to the correlation of layers 9 and 8 with layer X. Four of these fragments (16/116-159327 from the 2016–2022 excavation; R10874, R10879 and R10396 from the 1930s collection) also produced statistically indistinguishable radiocarbon dates (Fig. 2). The morphology and stable isotopic values20 of R10874 suggest that it originates from a different individual, consistent with a maternal relation. Notably, whereas nine of the ten reconstructed mtDNA genomes belonged to haplogroup N, one (16/116-159199) was identified as belonging to haplogroup R. When placed onto a phylogenetic tree with other ancient humans, the mtDNA genomes with an N haplogroup cluster together with the mtDNA genome of Zlatý kůň, an individual from the Czech Republic, whose chronological age is around 45,000 years before present on the basis of genetic estimates3 (Fig. 3 and Supplementary Figs. 26 and 27). The estimated mean genetic dates of the Ranis mtDNA genomes ranged from 49,105 to 40,918 years before present (Supplementary Table 22 and Supplementary Fig. 26), consistent with the radiocarbon dates from layers 9 and 8.
 www.nature.com
www.nature.com
The Middle to Upper Palaeolithic transition in Europe is associated with the regional disappearance of Neanderthals and the spread of Homo sapiens. Late Neanderthals persisted in western Europe several millennia after the occurrence of H. sapiens in eastern Europe1. Local hybridization between the two groups occurred2, but not on all occasions3. Archaeological evidence also indicates the presence of several technocomplexes during this transition, complicating our understanding and the association of behavioural adaptations with specific hominin groups4. One such technocomplex for which the makers are unknown is the Lincombian–Ranisian–Jerzmanowician (LRJ), which has been described in northwestern and central Europe5,6,7,8. Here we present the morphological and proteomic taxonomic identification, mitochondrial DNA analysis and direct radiocarbon dating of human remains directly associated with an LRJ assemblage at the site Ilsenhöhle in Ranis (Germany). These human remains are among the earliest directly dated Upper Palaeolithic H. sapiens remains in Eurasia. We show that early H. sapiens associated with the LRJ were present in central and northwestern Europe long before the extinction of late Neanderthals in southwestern Europe. Our results strengthen the notion of a patchwork of distinct human populations and technocomplexes present in Europe during this transitional period.
We tested 11 of the hominin remains for the preservation of ancient mitochondrial DNA (mtDNA). Between 4,413 and 175,688 unique reads mapping to the human mtDNA reference genome were recovered per skeletal fragment. These mtDNA reads had elevated frequencies of cytosine (C)-to-thymine (T) substitutions (32.6% to 49.6% on the 5′ end and 19.0% to 47.9% on the 3′ end, respectively; Supplementary Figs. 14–24), which are indicative of ancient DNA. Positions shown to be informative for differentiating between H. sapiens, Neanderthal and Denisovan mtDNA genomes enabled us to identify each of the 11 skeletal fragments as belonging to ancient H. sapiens (Supplementary Table 18). Libraries from ten of the eleven skeletal fragments contained sufficient data for reconstructing near-complete mtDNA genomes. Five of these mtDNA genomes showed no pairwise differences among them for the positions covered, suggesting that they stemmed from either the same individual or maternally related individuals (Supplementary Fig. 25 and Supplementary Table 19). Four of these skeletal fragments come from the 1932–1938 collection and one from the 2016–2022 excavation (16/116-159327; Fig. 1), providing additional support to the correlation of layers 9 and 8 with layer X. Four of these fragments (16/116-159327 from the 2016–2022 excavation; R10874, R10879 and R10396 from the 1930s collection) also produced statistically indistinguishable radiocarbon dates (Fig. 2). The morphology and stable isotopic values20 of R10874 suggest that it originates from a different individual, consistent with a maternal relation. Notably, whereas nine of the ten reconstructed mtDNA genomes belonged to haplogroup N, one (16/116-159199) was identified as belonging to haplogroup R. When placed onto a phylogenetic tree with other ancient humans, the mtDNA genomes with an N haplogroup cluster together with the mtDNA genome of Zlatý kůň, an individual from the Czech Republic, whose chronological age is around 45,000 years before present on the basis of genetic estimates3 (Fig. 3 and Supplementary Figs. 26 and 27). The estimated mean genetic dates of the Ranis mtDNA genomes ranged from 49,105 to 40,918 years before present (Supplementary Table 22 and Supplementary Fig. 26), consistent with the radiocarbon dates from layers 9 and 8.

Homo sapiens reached the higher latitudes of Europe by 45,000 years ago - Nature
Through archaeological excavation, morphological and proteomic taxonomic identification, mitochondrial DNA analysis and direct radiocarbon dating of human remains, a study reports the presence of Homo sapiens in Germany north of the Alps more than 45,000 years ago.
Last edited:

