norbert
Regular Member
I have come across the following paper from 2014 before and stumbled upon it again today by chance: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0105090#pone-0105090-g002
First off, a number of historical inaccuracies occur throughout the text. We learn that the Illyrians were assimilated by the Slavs while those who weren't, probably fled to the south, to present-day Albania. I suppose the authors (17 of them!) had no idea that this is the area where the ancient Greek encountered the Illyrians in the first place. The Illyrian name itself most likely belonged to a single tribe which the Greek then applied to other tribes further north who may have shared similar customs and language. It is also mentioned that "Mongolian tribes" moved to the Balkans. Really? Turns out, those "Mongolian tribes" are Huns, Avars and Turks. They were Mongolian? Whatever they were, when exactly did they move to the Balkans? The Avars were in Pannonia but they were a minority warrior caste that pretty much disappeared.
Regarding samples, the authors say
"Genome-wide autosomal markers of 70 Western Balkan individuals from Bosnia and Herzegovina, Serbia, Montenegro, Kosovo and former Yugoslav Republic of Macedonia (see map in Figure 1) together with the published autosomal data of 20 Croatians were analyzed in the context of 695 samples of global range (see details from Table S1)."
Seriously? 70 samples for the whole Western Balkans? Now look at the chart from this publication:
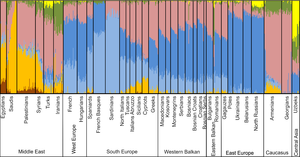
It doesn't make any sense. Take the South Slavs. 4 Bosnian Serb samples are supposed to tell us something about the autosomal ancestry of the Bosnian Serbs in general? What about the "Bosniacs" (I seem to recall it's BosniaKs)? Notice how they have slightly elevated "Middle Eastern" results compared to Serbs and Croats? There are 15 Bosniak samples. Swap them with another 15 and you'll get completely different results. Same goes for all the other samples, of course.
Then there is the problem with the categories used here:
"ADMIXTURE analysis of autosomal SNPs of the Balkan region in a global context on the resolution level of 7 assumed ancestral populations: the African (brown), South/West European (light blue), Asian (light yellow), South Asian (Green), Middle Eastern (dark yellow), North/East European (dark blue) and beige Caucasus component."
What kind of category is South/West European? What do they mean by Middle Eastern? Or South Asian? How can France have so much "South Asian" as shown in the chart? These are hardly scientific categories. I guess if they featured the Finns in the chart, they'd be like 60% "East Asian" and the Swedes probably to a third. What is East Asian? I get the impression that these interpretations are based on Y-DNA haplogroups despite the autosomal claim but even that doesn't make sense. Is everyone who belongs to haplogroup N1c East Asian? They also lump together North and sub-Saharan Africans. That's like putting together Indians and the Chinese.
You look at 2 papers and you get 4 different results and even more interpretations. In academia it is very important to publish as much as possible and this goes for all scientific professions. I suppose amateurs can afford to fool around with their software "oracles" but serious scientists have a responsibility to details and accuracy.
First off, a number of historical inaccuracies occur throughout the text. We learn that the Illyrians were assimilated by the Slavs while those who weren't, probably fled to the south, to present-day Albania. I suppose the authors (17 of them!) had no idea that this is the area where the ancient Greek encountered the Illyrians in the first place. The Illyrian name itself most likely belonged to a single tribe which the Greek then applied to other tribes further north who may have shared similar customs and language. It is also mentioned that "Mongolian tribes" moved to the Balkans. Really? Turns out, those "Mongolian tribes" are Huns, Avars and Turks. They were Mongolian? Whatever they were, when exactly did they move to the Balkans? The Avars were in Pannonia but they were a minority warrior caste that pretty much disappeared.
Regarding samples, the authors say
"Genome-wide autosomal markers of 70 Western Balkan individuals from Bosnia and Herzegovina, Serbia, Montenegro, Kosovo and former Yugoslav Republic of Macedonia (see map in Figure 1) together with the published autosomal data of 20 Croatians were analyzed in the context of 695 samples of global range (see details from Table S1)."
Seriously? 70 samples for the whole Western Balkans? Now look at the chart from this publication:

It doesn't make any sense. Take the South Slavs. 4 Bosnian Serb samples are supposed to tell us something about the autosomal ancestry of the Bosnian Serbs in general? What about the "Bosniacs" (I seem to recall it's BosniaKs)? Notice how they have slightly elevated "Middle Eastern" results compared to Serbs and Croats? There are 15 Bosniak samples. Swap them with another 15 and you'll get completely different results. Same goes for all the other samples, of course.
Then there is the problem with the categories used here:
"ADMIXTURE analysis of autosomal SNPs of the Balkan region in a global context on the resolution level of 7 assumed ancestral populations: the African (brown), South/West European (light blue), Asian (light yellow), South Asian (Green), Middle Eastern (dark yellow), North/East European (dark blue) and beige Caucasus component."
What kind of category is South/West European? What do they mean by Middle Eastern? Or South Asian? How can France have so much "South Asian" as shown in the chart? These are hardly scientific categories. I guess if they featured the Finns in the chart, they'd be like 60% "East Asian" and the Swedes probably to a third. What is East Asian? I get the impression that these interpretations are based on Y-DNA haplogroups despite the autosomal claim but even that doesn't make sense. Is everyone who belongs to haplogroup N1c East Asian? They also lump together North and sub-Saharan Africans. That's like putting together Indians and the Chinese.
You look at 2 papers and you get 4 different results and even more interpretations. In academia it is very important to publish as much as possible and this goes for all scientific professions. I suppose amateurs can afford to fool around with their software "oracles" but serious scientists have a responsibility to details and accuracy.

