I1a3_Young
Regular Member
- Messages
- 550
- Reaction score
- 60
- Points
- 28
- Location
- FL
- Ethnic group
- Basically British
- Y-DNA haplogroup
- I1 Z63*
- mtDNA haplogroup
- H5b1
Credit to the GeneticGeneologist:
http://www.geneticgenealogist.net/2016/01/how-to-get-ydna-haplogroup-from.html
The Ancestry.com raw data file (which will be emailed to you after you log in and request it) contains 1691 lines of Y-DNA data.
1. Open the text file in Excel and edit it to match the 23andMe raw data output. Remove all the language at the top, delete "allele 2" column, rename "allele 1" to "genotype", find and replace all "24" with "Y". You only need the 1691 rows with Y data, not all the rest from the raw data file.
2. Download and run 23andMe To YSNPs.exe (from link above) which will create a text file of SNP matches. You can manually do it someplace like the ISOGG Y-browser but it would take forever. I recalculated a few to satisfy my curiosity.
3. Use the Morley Y-DNA Haplogroup predictor (copy/paste the results from the created SNP text file) https://ytree.morleydna.com/
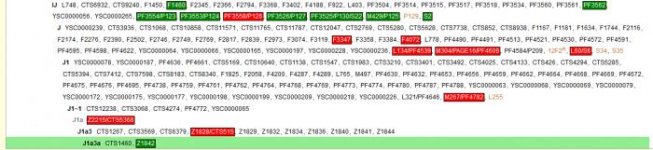
You get visual trees of positive and negative matches and a "most likely" suggestion. You should be able to see the many green matches in your tree, for example I show many "green" indicators in C, F, IJ, I, and I1. Then one match for I1a3 and red for the other I1a branches. I got a single positive for a "J3a something" but since no branches "above" in the J line are matches, I can ignore it.
I have sent off my LivingDNA kit so I will compare the results when they arrive. Does anyone have an Ancestry test to compare to another Y-DNA test so we can test the accuracy?
http://www.geneticgenealogist.net/2016/01/how-to-get-ydna-haplogroup-from.html
The Ancestry.com raw data file (which will be emailed to you after you log in and request it) contains 1691 lines of Y-DNA data.
1. Open the text file in Excel and edit it to match the 23andMe raw data output. Remove all the language at the top, delete "allele 2" column, rename "allele 1" to "genotype", find and replace all "24" with "Y". You only need the 1691 rows with Y data, not all the rest from the raw data file.
2. Download and run 23andMe To YSNPs.exe (from link above) which will create a text file of SNP matches. You can manually do it someplace like the ISOGG Y-browser but it would take forever. I recalculated a few to satisfy my curiosity.
3. Use the Morley Y-DNA Haplogroup predictor (copy/paste the results from the created SNP text file) https://ytree.morleydna.com/
You get visual trees of positive and negative matches and a "most likely" suggestion. You should be able to see the many green matches in your tree, for example I show many "green" indicators in C, F, IJ, I, and I1. Then one match for I1a3 and red for the other I1a branches. I got a single positive for a "J3a something" but since no branches "above" in the J line are matches, I can ignore it.
I have sent off my LivingDNA kit so I will compare the results when they arrive. Does anyone have an Ancestry test to compare to another Y-DNA test so we can test the accuracy?