Jovialis
Advisor
- Messages
- 9,313
- Reaction score
- 5,876
- Points
- 113
- Ethnic group
- Italian
- Y-DNA haplogroup
- R-PF7566 (R-Y227216)
- mtDNA haplogroup
- H6a1b7
Abstract
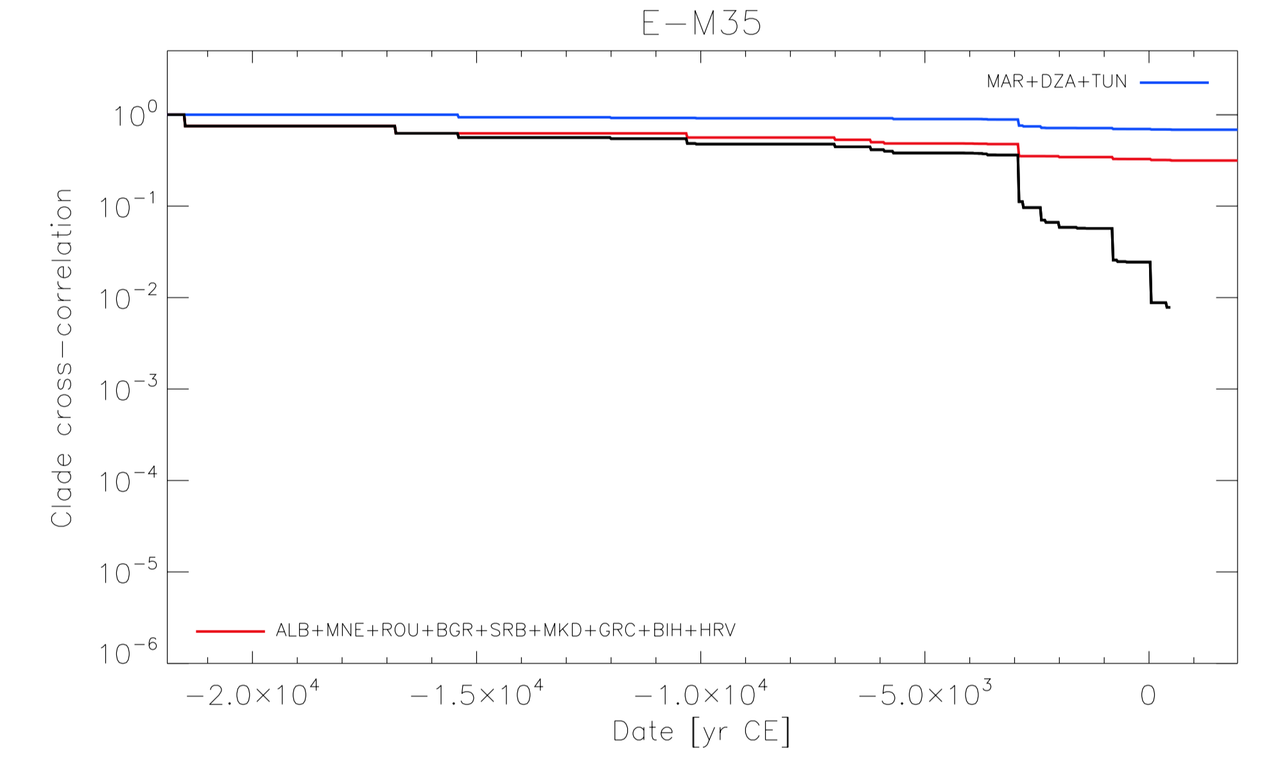
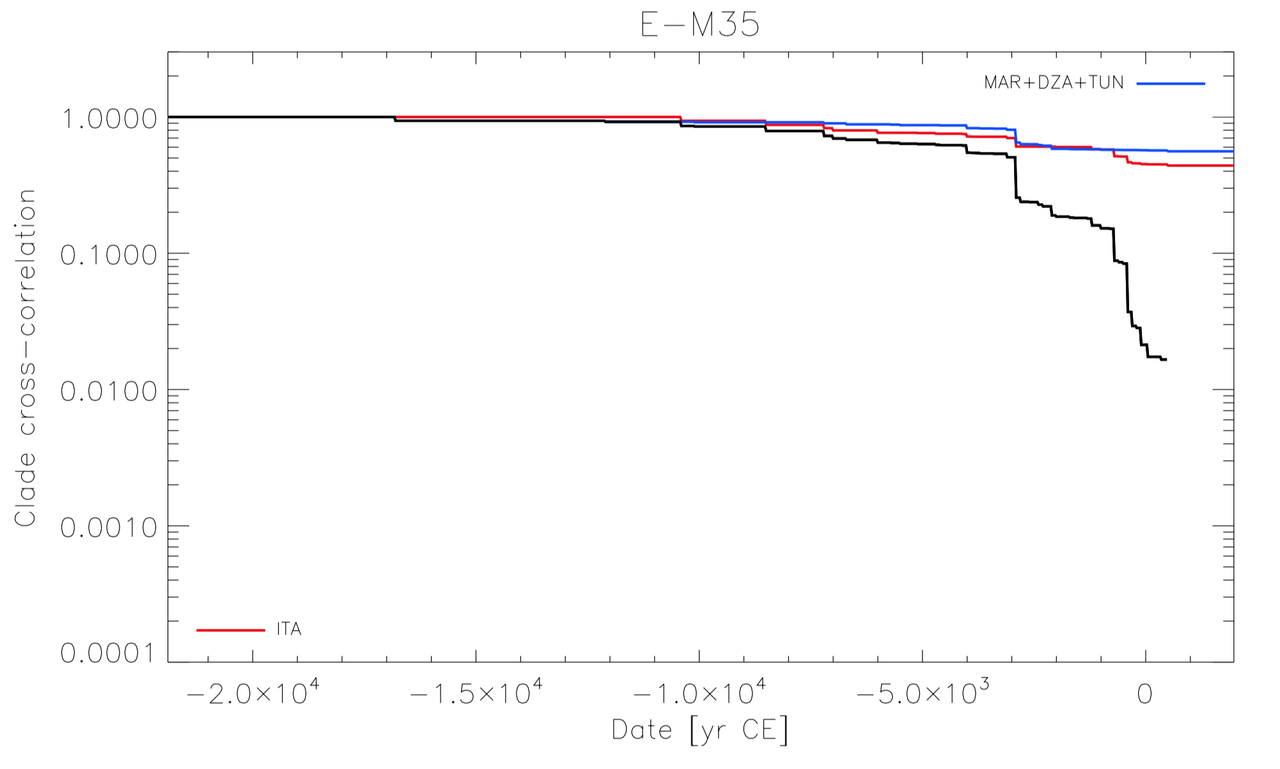
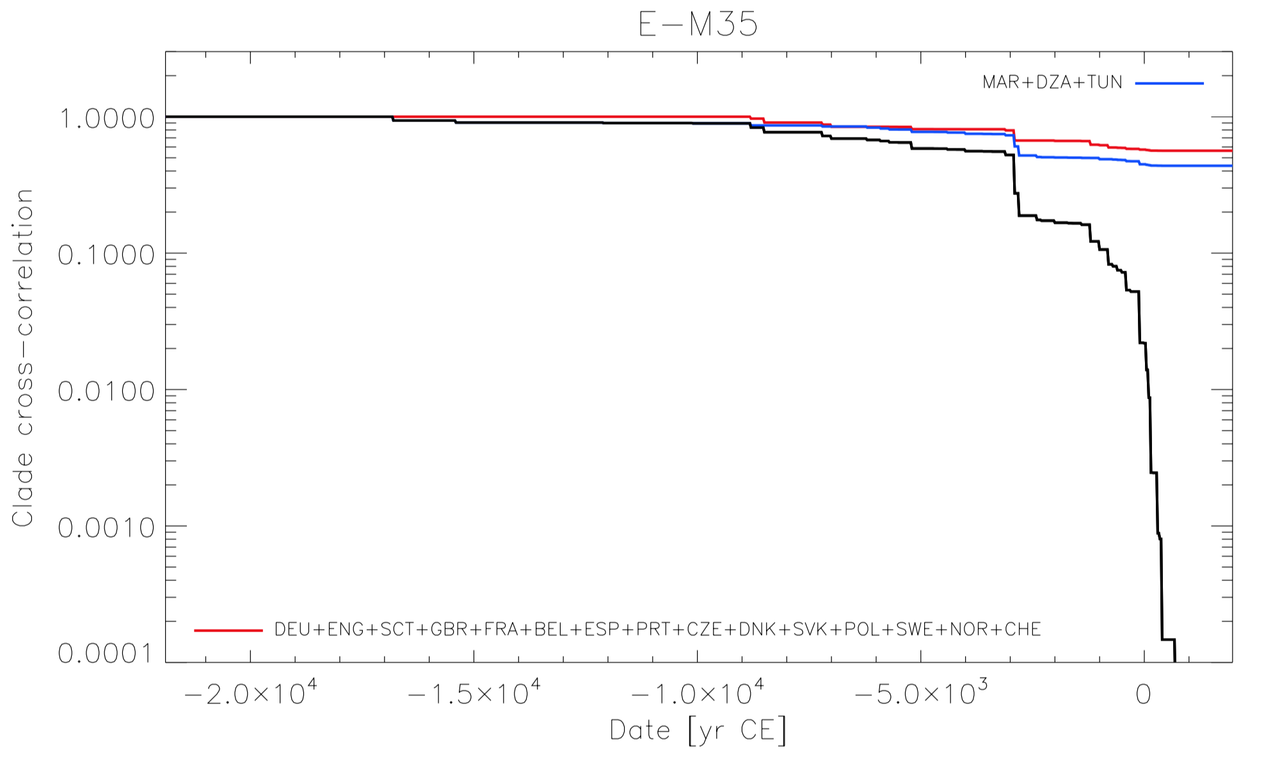
Ancient DNA research in the past decade has revealed that European population structure changed dramatically in the prehistoric period (14,000–3000 years before present, YBP), reflecting the widespread introduction of Neolithic farmer and Bronze Age Steppe ancestries. However, little is known about how population structure changed from the historical period onward (3000 YBP - present). To address this, we collected whole genomes from 204 individuals from Europe and the Mediterranean, many of which are the first historical period genomes from their region (e.g. Armenia and France). We found that most regions show remarkable inter-individual heterogeneity. At least 7% of historical individuals carry ancestry uncommon in the region where they were sampled, some indicating cross-Mediterranean contacts. Despite this high level of mobility, overall population structure across western Eurasia is relatively stable through the historical period up to the present, mirroring geography. We show that, under standard population genetics models with local panmixia, the observed level of dispersal would lead to a collapse of population structure. Persistent population structure thus suggests a lower effective migration rate than indicated by the observed dispersal. We hypothesize that this phenomenon can be explained by extensive transient dispersal arising from drastically improved transportation networks and the Roman Empire’s mobilization of people for trade, labor, and military. This work highlights the utility of ancient DNA in elucidating finer scale human population dynamics in recent history.Stable population structure in Europe since the Iron Age, despite high mobility
Reconstructing human mobility patterns using historical period genomes illustrates how the Roman Empire’s military and economic activities catalyzed an era of transient movement against the backdrop of generations-long prehistoric migrations.