I think you mean Z38240. I've seen you made this typo before, so just wanted to point it out.
Yes indeed, thanks for pointing it.
I'm always messing around with "Z"s and "Y"s subclades name ... and I wrote my last message very quickly, without a single re-reading.
I believe your theories will not hold once we get some relevant aDNA for these subclades. We can't rely on modern distribution and YFull TMRCA estimates (which merely tell us when the lineages split, not when migrations occured)
I think, I already point this out, but basically, to create some degree of diversity you need a minimum level of migration, or at least some "diffusion".
If not, lineage are screening each other and you cannot build significant surviving diversity. Basically, an explosion of diversity == migration/expansion of the carriers.
It is hard to migrate without creating diversity, the migration need to be very quick to not let significant clades behind, or in a population poorly interacting with others during their migration.
Mobility create diversity (that will be convolved by later migrations or partially erased by later invasions).
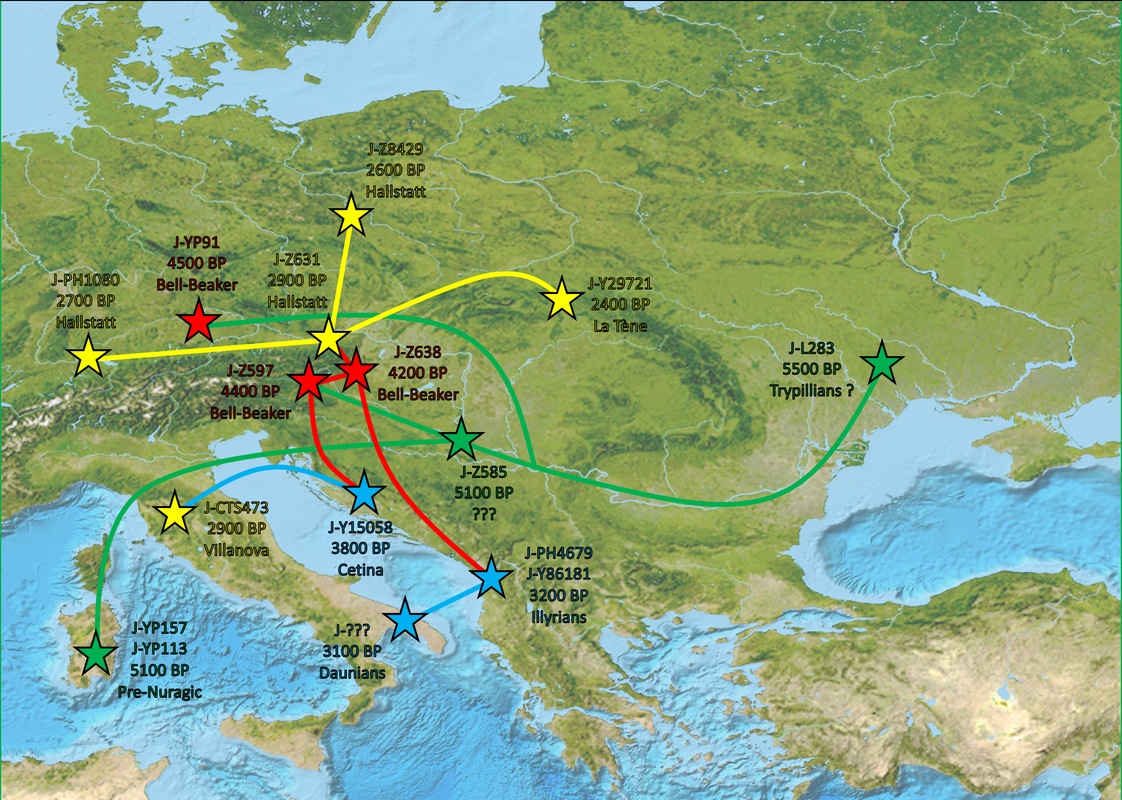
and assume things were the same over 2000 ybp, and on top of that ignore ancient DNA! We have a ~J-Z1297* dated ~1450 BC, (where btw J-Y23094, J-Y21878, J-Z631 stem from) just north of Albania while lacking elsewhere.
And you have Z2507- in the north.
We already made this disucussion. You find this clade, cousin to Y15058 in an area where a clear Y15058 diffusion occured between J-Z1297 definition-time and the sample lifetime.
Then, this sample is not very indicative, like the Z2507-. Because we know of a convolution event that can explain both samples either with a north-south or a south-north migration.
Then, you should apply to this samples the same logic you use for discarding modern diversity. And then, you can't consider this sample because it is too "recent" compared to what you want to probe. If it was in the range 2200-1800bc it would have been more relevent.
You have two big things when you play with data :
--> Transfert function
--> Statistical noise
Modern data have a significantly better statistical noise, but a more complexe transfert function (more migrations/invasions).
Ancient DNA have a very poor Statistical noise, making it barely relevent in area where the population was significantly moving when you let pass few hundreds of years (because transfert function is already very complicated).
I know what I chose, because I didn't know a single case where diversity fails completely.
I think it was you who mentioned some R1b clades weird diversity centers, some L283-diversity center in Americas ... none of that exists.
It only shows that you don't really know how a proper diversity analysis works.
Which is not a big deal, but then it is weird to see you discarding such approach when you obviously didn't understand well this methodology.
So logic would say there was likely more than just J-PH4679 in the region, likely including J-Y23094 lineages among others.
Not really, maybe this is the conclusion you like. But diversity is not going there. And ancient DNA didn't support such conclusion, especially with one single sample, very late, compared to the clade diversification, and in an area were cousin clades diffused.
With regards to J-PH4679, it's not as "exclusive" to Albania as the YFull tree suggests. There is actually already an American sample (likely western European origin) who splits this subclade, as he is showing positive for PH2967 and negative for PH4679 itself.
PH2967+ ? Thus he is Z21045, or nearly Z21045. But PH4679- ... thus this sample is not PH4679.
Good point, it match exactly with what I'm saying.
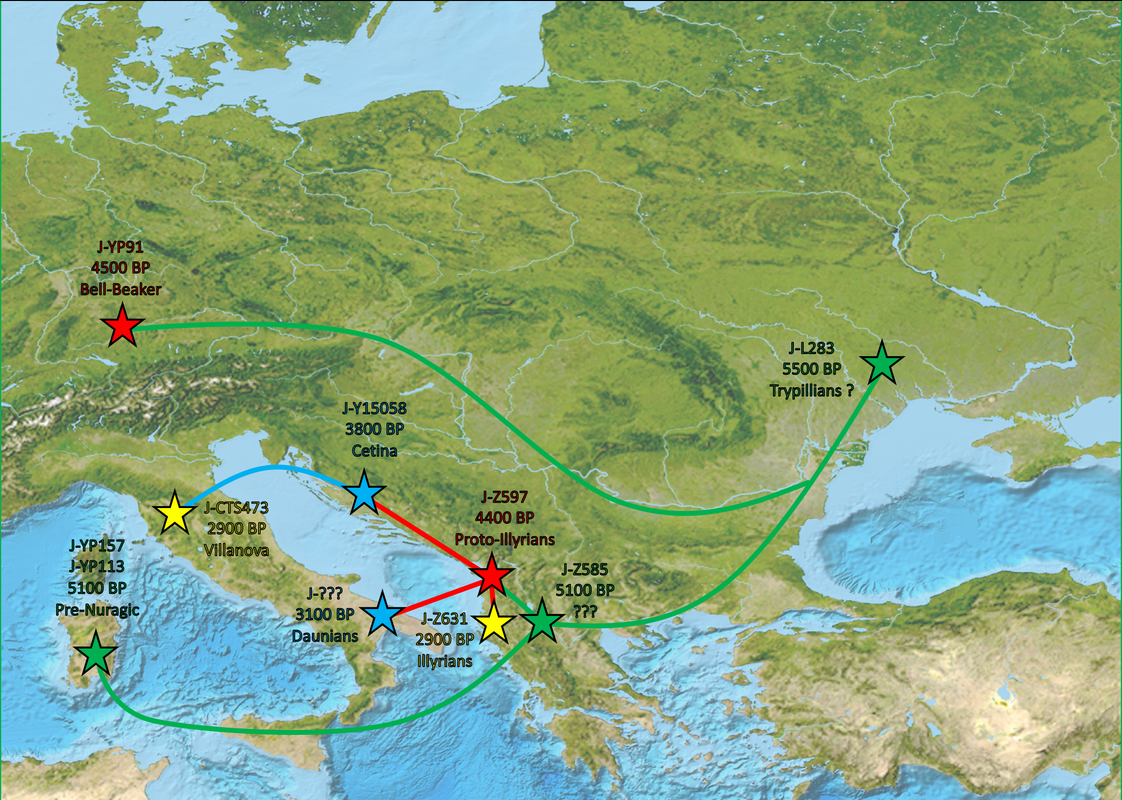
I would place Z21045 either around north Italy or Slovenia by ~1800 BC, then the branch that will give PH4679, migrated south with the 1800-1700 BC expansion on the Adriatic coast, and started diversification way later around 1200 BC in southern Illyria.
Thus, I'm not surprised that some PH2967 will be found in western Europe if digging a bit more. Be sure there is some in France ... maybe me, who knows ? (Yes because my Z631 call is a two-SNPs* based argument ... it is not super robust yet).
*Using a two-SNPs based calls, I would also be I2a1 ... showing how a two-SNP based haplogroup can be completely wrong.
There is also a sample with Italian paternal origin who is SNP tested Z1296+, Z1297- and STR signature suggests he probably splits J-PH4679>Z38300>Y20899. But of course, you won't see these samples on YFull (at least not yet).
I discuss, and perform reasoning, with data I can consults ... not with rumored samples.
But ok, let speak about this case :
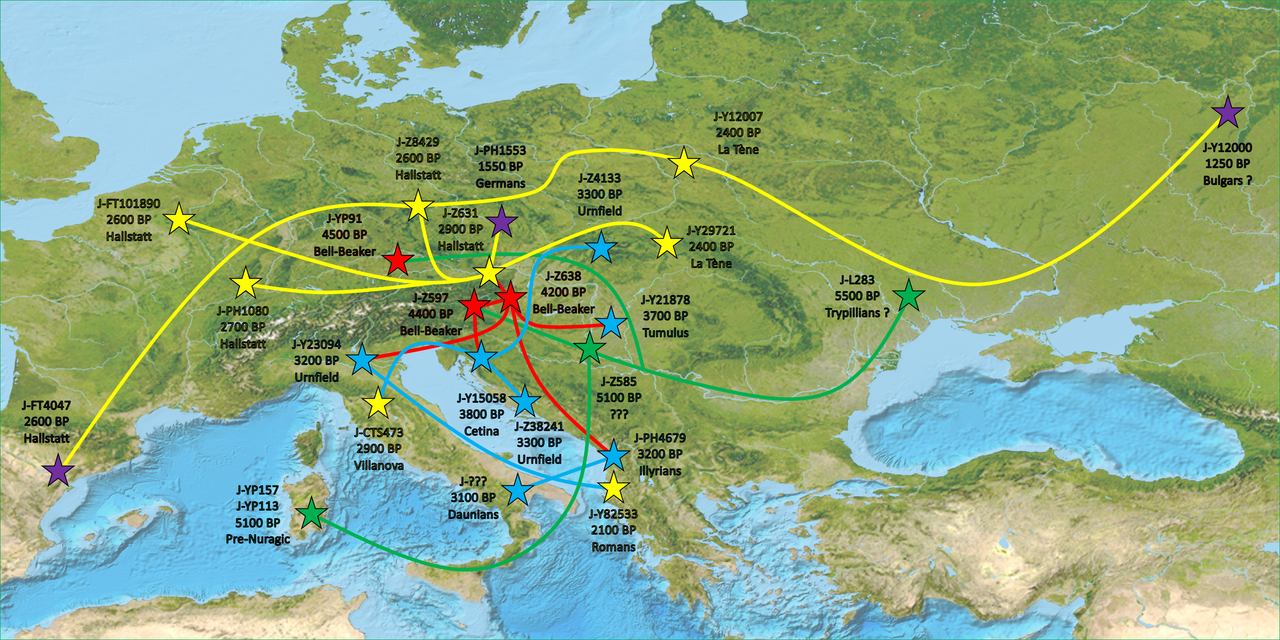
Y20899 ? Ok, then I would go for a consequence of invasion of Illyria by romans, diversification is in fact right at the good moment ~2100 BP.
But anyway, it is not a problem even for a deeper subclade. Albanians are migrating humans too, I don't understand the point you try to make here ?
Could Albanians cross the Adriatic to enter Italy ? For sure they can. And nothing avoid such thing to happen between 3200 bp and modern day.
Unless you trace the paternal origin deep enough to enter the range of the haplogroup definition-times ... I don't see how this exemple is relevent.
But some clades seems to show that clades are having hard time to exit Albania. It is related to the shape of the philogenic tree. Once in Albania, we don't see many subclades exiting ... whereas, when a clade appear in Alabania, it is embeded in a bunch of non-Albanian subclades.
This is a pattern indicating a migration toward Albania, not from Albania.
So by this logic, you can make the same argument, that J-PH4679 was either spread by "Urnfield" and related movements or "injected to Albania" in the Roman times.
Not working that way.
PH4679 --> Two pre-roman diversity event with Albanian samples. Plus, ancient DNA did in fact proves some Z638 went there by ~1500 BC.
Y82533 --> no Albanian diversity prior 2100 BP. Precedent diversity is in Italy in 1200 BP.
A J-L283 expansion in the adriatic coast occured around 1800-1700 BC (even if for some reasons it appear that you deny that), and Y86181 was not among them.
First moment we find trace of these guys is after Roman conquest, and before that the last diversity point we have is Italy. Unless going for a migration from Italy --> Albania in the range 3200-2100 BP, it sounds more reasonable to go for an arrival date around 2100 BP with the romans.
It might be proven wrong, but yet, it is the more likely.
I think people on this topic are losing from their perspective that L283 is not an Illyrian clade, Illyrian wasn't a thing in 5500 BP.
There is not reasons to belive that L283 travelled as a pack for 2000+ years before becoming Illyrians ! To be correct, such model didn't work (because lineage screening considerations).
Even if some L283 are Illyrians (and Patterson et al. 2021 proved it with a crazy high fraction of J-L283 on the adriatic coast), it didn't means that all L283 that you will found in Albania will be Illyrian lineages.
You get the idea. Pretty much all these J-L283 subclades show similar distribution: Regional subclade diversity in the western Balkans (J-Z38240 subclades more northern, while J-Z638 subclades more common among Albanians), and then isolated lineages of these subclades throughout Europe with usually no deep regional subclade diversity which would point to no regional continuity, but likely a combination of different migrations (including the Roman period).
Weird diversity analysis. We are not at all converging to the same thing.
L283 diversity is fairly homogeneous on the European continent (excluding France, Spain, and Portugal ... but they are poorly sampled countries, when better sampled their diversity will rise Again ... some YP91 are probably lurking around).
Albania didn't show strong diversity despite having a giant oversampling bias.
How do you perform diversity analyses ? Looking at Flags on Y-Full ?
In fact Z638 shows central European diversity (~Slovenia/Austria/Hungary/Slovaquia).
You find a lot of Albanian in some clades because Albania is over-sampled in Y-Full (it is a well known bias).
In non-oversampled data for Albania (FTDNA for instance), diversity in Albania is very small.
With data oversampled in UK, you manage to find diversity center for Z631 in UK ... In this case too, it is simply over-sampling bias.
In fact, what clades distribution in Albania shows is that it received a large number of contribution by L283 peoples through multiple events.
Again, remove PH4679, and your L283 fraction become the same as all other countries. The L283 clade distribution is already similar to all other country.
Therefore, it means that PH4679 has something to do with history of Albanian lands ... but that likely, the history of other clades in Albania is just an history shared by Europeans. Then each clade needs to be considered independely to identify when and how they enter Albanian land.
But not all clades share the same solution.
One should also study and understand Balkans history and put things into context. Ex: The Illyrians were the first in the Balkans to be conquered by the Romans. Later, during late antiquity/early medieval migrations, the Slavic tribes heavily settled these regions, which would contribute towards a "replacement and displacement" of Y-lineages that were there before.
And some J-L283 lineage in Albania were probably brought there by slavic peoples ... there is few very good candidates. But I won't enter this discussion too deeply, I know too well where it goes.
G.