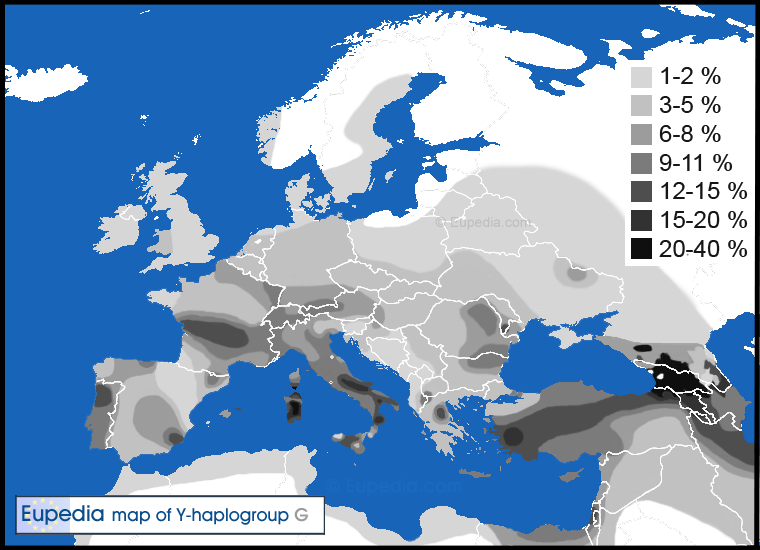
Geographic distribution
Nowadays haplogroup G is found all the way from Western Europe and Northwest Africa to Central Asia, India and East Africa, although everywhere at low frequencies (generally between 1 and 10% of the population). The only exceptions are the Caucasus region, central and southern Italy and Sardinia, where frequencies typically range from 15% to 30% of male lineages.
The overwhelming majority of Europeans belong to the G2a subclade, and most northern and western Europeans fall more specifically within G2a-L140 (or to a lower extend G2a-M406). Almost all G2b (L72+, formerly G2c) found in Europe are Ashkenazi Jews. G2b is found from the Middle East to Pakistan, and is almost certainly an offshoot of Neolithic farmers from western Iran, where G2b was identified in a 9,250 year-old sample by Broushaki et al. (2016).
Haplogroup G1 is found predominantly in Iran, but is also found in the Levant, among Ashkenazi Jews, and in Central Asia (notably in Kazakhstan).
G2a makes up 5 to 10% of the population of Mediterranean Europe, but is relatively rare in northern Europe. The only regions where haplogroup G2 exceeds 10% of the population in Europe are in Cantabria in northern Spain, in northern Portugal, in central and southern Italy (especially in the Apennines), in Sardinia, in northern Greece (Thessaly), in Crete, and among the Gagauzes of Moldova - all mountainous and relatively isolated regions. Other regions with frequencies approaching the 10% include Asturias in northern Spain, Auvergne in central France, Switzerland, Sicily, the Aegean Islands, and Cyprus.
Distribution of haplogroup G in Europe, North Africa and the Middle East
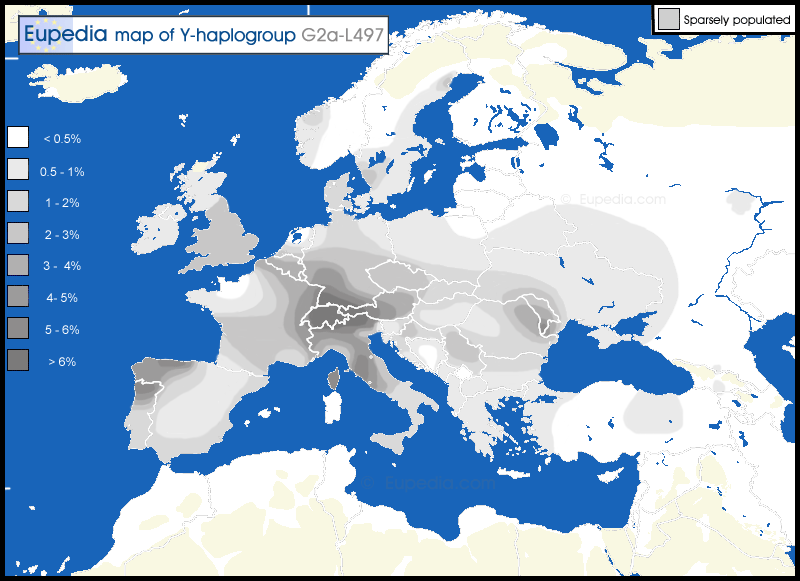
Distribution map of haplogroup G2a-L497 in Europe
Distribution map of haplogroup G2a-U1 in Europe

Distribution map of haplogroup G2a-M406 in Europe

Distribution map of haplogroup G2a-L293 in Europe

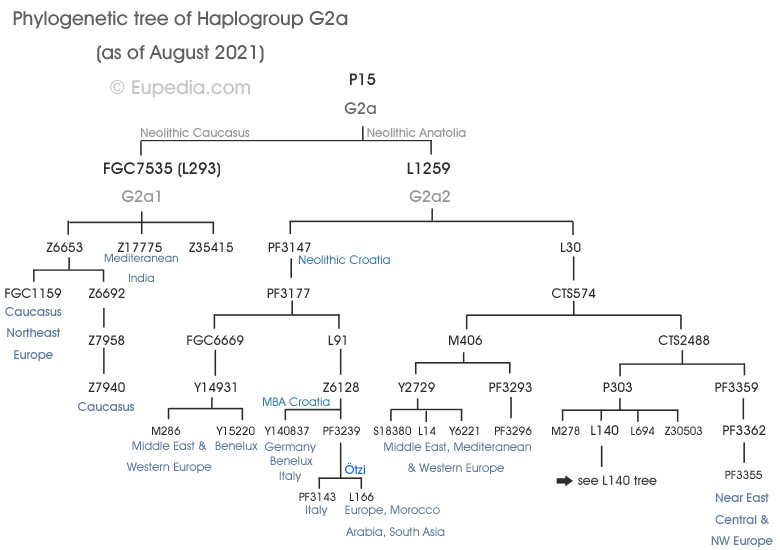
Phylogeny of G2a
If you are new to genetic genealogy, please check our Introduction to phylogenetics to understand how to read a phylogenetic tree.
Origins
Haplogroup G descends from macro-haplogroup F, which is thought to represent the second major migration of Homo sapiens out of Africa, at least 60,000 years ago. While the earlier migration of haplogroups C and D had followed the coasts of South Asia as far as Oceania and the Far East, haplogroup F penetrated through the Arabian peninsula and settled in the Middle East. Its main branch, macro-haplogroup IJK would become the ancestor of 80% of modern Eurasian people. Haplogroup G formed approximately 50,000 years ago as a side lineage of haplogroup IJK, but seems to have had a slow start, evolving in isolation for tens of thousands of years, possibly in the Near East, cut off from the wave of colonisation of Eurasia.
As of late 2016, there were 303 mutations (SNPs) defining haplogroup G, confirming that this paternal lineage experienced a severe bottleneck before splitting into haplogroups G1 and G2. G1 might have originated around modern Iran at the start of the Last Glacial Maximum (LGM), some 26,000 years ago. G2 would have developed around the same time in West Asia. At that time humans would all have been hunter-gatherers, and in most cases living in small nomadic or semi-nomadic tribes. Members of haplogroup G2 appear to have been closely linked to the development of early agriculture in the Fertile Crescent part, starting 11,500 years before present. The G2a branch expanded to Anatolia, the Caucasus and Europe, while G2b diffused from Iran across the Fertile Crescent and east to Pakistan. It is now found mostly among Lebanese and Jewish people, but also at low frequency in the Arabian peninsula, Syria, Iraq, Iran, Afghanistan and Pakistan.
There has so far been ancient Y-DNA analysis from Early Neolithic Anatolia, Iran, Israel, Jordan as well as most Neolithic cultures in Europe (Thessalian Neolithic in Greece, Starčevo culture in Hungary/Croatia, LBK culture in Germany, Remedello in Italy, and Cardium Pottery in south-west France and Spain) and all sites yielded a majority of G2a individuals, except those from the Levant. This strongly suggests that farming was disseminated by members of haplogroup G at least from Anatolia/Iran to Europe. Lazaridis et al. (2016) tested 44 ancient Near Eastern samples, including Neolithic farmers from Jordan and western Iran, and found one G2b sample dating from the Pre-Pottery Neolithic (c. 7,250 BCE) and a G2a1 from the Early Pottery Neolithic (c. 5,700 BCE), both from Iran. The few samples from the Levant belonged to haplogroups CT, E1b, H2 and T, but it cannot be ruled out yet that haplogroup G will show up in other samples. Mathieson et al. (2015) tested the Y-DNA of 13 Early Neolithic farmers from the Barcın site (6500-6200 BCE) in north-western Anatolia, and 8 of them belonged to haplogroup G2a (subclades G2a2a-PF3146, G2a2a1b-L91, G2a2a1b1-PF3247, G2a2b-L30, G2a2b2a-P303, G2a2b2a1c-CTS342). The other samples belonged to haplogroups C1a2, H2, I, I2c and J2a. These minor haplogroups also show up among Early Neolithic farmers in the Balkans, once again amongst a G2a majority. Occasionally other Near Eastern lineages showed up, like one T1a sample in the LBK culture, and one R1b-V88 in northwest Spain. T1a tribes are thought to have domesticated ovicaprids in the Zagros mountains, while R1b tribes would have domesticated cattle in the north of the Fertile Crescent.
The highest genetic diversity within haplogroup G is found in the northern part of the Fertile Crescent, between the Levant and the Caucasus, which is a good indicator of its region of origin. It is thought that early Neolithic farmers expanded from northern Mesopotamia westwards to Anatolia and Europe, eastwards to South Asia, and southwards to the Arabian peninsula and North and East Africa. So far, the only G2a people negative for subclades downstream of P15 or L149.1 were found exclusively in the South Caucasus region.
History of haplogroup G2a
Several historical migrations brought different subclades of haplogroup G to Europe or redistributed them geographically.
Neolithic farmers and mountain herders
The testing of Neolithic remains in various parts of Europe has confirmed that haplogroup G2a was the dominant lineages of Neolithic farmers and herders who migrated from Anatolia to Europe between 9,000 and 6,000 years ago.
Cereal and legume farming first developed 11,500 years ago in the Fertile Crescent, in what is now Israel/Palestine, Jordan, Lebanon, Syria and Iraq, but did not expand much beyond this region for the first two and a half millennia. The reason for this delay was that early agriculture was too rudimentary to allow an independent subsistence and was merely a way of supplementing the diet of hunter-gatherers. Cultivation started with wheat, figs and legumes. The domestication of wheat and barley was a lengthy process that necessitated the selection of cultivars that possess mutations for larger, less brittle and nonshattering spikes. The flood plains of Mesopotamia were ideal for primitive cereal farming as they did not require irrigation.
Pottery first appears in the Near East approximately 9,000 years ago in northern Mesopotamia. The development of pottery seems to coincide with the sudden expansion of G2a agriculturalists toward western Anatolia and Europe. Pottery allowed easy storing of cereals and legumes and could have facilitated trade with neighbouring ovicaprid and cattle herders, and pig farmers. Goats and sheep had first been domesticated some 11,000 years ago in the Zagros and Taurus mountains on the northern edge of the Fertile Crescent, but were not introduced to the Levant until approximately 8,500 years ago (see The development of goat and sheep herding during the Levantine Neolithic, A. Wasse, pp. 26-27), just after the appearance pottery.
The Neolithic settlement of Çatalhöyük in south-central Anatolia was founded by cereal and pulse farmers who also brought domesticated goats and sheep. Only a few centuries later (c. 6500 BCE) were cattle introduced to Çatalhöyük and other sites in Central Anatolia, presumably by trading with their eastern neighbours. Also around 8,500 years ago, G2a Neolithic farmers arrived in northwest Anatolia and Thessaly in central Greece, as attested by the ancient genomes sequenced by Mathieson et al. (2015) and Hofmanová et al. (2015). G2a farmers from the Thessalian Neolithic quickly expanded across the Balkans and the Danubian basin, reaching Serbia, Hungary and Romania by 5800 BCE, Germany by 5500 BCE, and Belgium and northern France by 5200 BCE. Ancient skeletons from the StarčevoKőrösCriș culture (6000-4500 BCE) in Hungary and Croatia, and the Linear Pottery culture (5500-4500 BCE) in Hungary and Germany, all confirmed that G2a (both G2a2a and G2a2b) remained the principal paternal lineage even after farmers intermingled with indigenous populations as they advanced. Freilich et al. (2021) analysed seven samples from the Middle Neolithic Sopot culture in northeastern Croatia, three of which belonged to G2a (two G-PF3147 and one G-U1>L13>Z2022).
By 7,800 years ago, farmers making cardial pottery arrived at the Marmara coast in northwest Anatolia with ovicaprids and pigs. These people crossed the Aegean by boat and colonized the Italian peninsula, the Illyrian coast, southern France and Iberia, where they established the Cardium Pottery culture (5000-1500 BCE). Once again, ancient DNA yielded a majority of G2a samples in the Cardium Pottery culture, with G2a frequencies above 80% (against 50% in Central and Southeast Europe).
Nevertheless, substantial minorities of other haplogroups have been found on different Neolithic sites next to a G2a majority, including C1a2, H2, I*, I2a1, I2c, and J2a in Anatolia, C1a2, E-M78, H2, I*, I1, I2a, I2a1, J2 and T1a in Southeast and Central Europe (Starčevo, Sopot, LBK), as well as E-V13, H2, I2a1, I2a2a1 and R1b-V88 in western Europe (Cardium Pottery, Megalithic). H2 and T1a were found in the Pre-Pottery Neolithic Levant and are undeniably linked to the early development of agriculture alongside G2a. That being said, C1a2 was also found in Mesolithic Spain (Olalde et al. 2014) and, as it is an extremely old lineage associated with the first Paleolithic Europeans, it could have been found all over Europe and Anatolia before the Neolithic. E1b1b was also found in the Pre-Pottery Neolithic Levant, but the subclades may not be E-M78 or E-V13 (more likely E1b1b1* or E-M123). R1b-V88 surely spread from the Near East too, although through a different route, with cattle herders via North Africa, then crossing over to Iberia. The rest probably represent assimilated hunter-gatherers descended from Mesolithic western Anatolian (I*, I2c, J2) and Europeans (E-V13, I*, I1, I2a, I2a1, I2a2). It is interesting to note that many of these lineages, such as C1a2, H2 and I* are virtually extinct anywhere nowadays, and several others are now very rare in Europe (I2c, R1b-V88).
Expansion of agriculture from the Middle East to Europe (9500-3800 BCE)
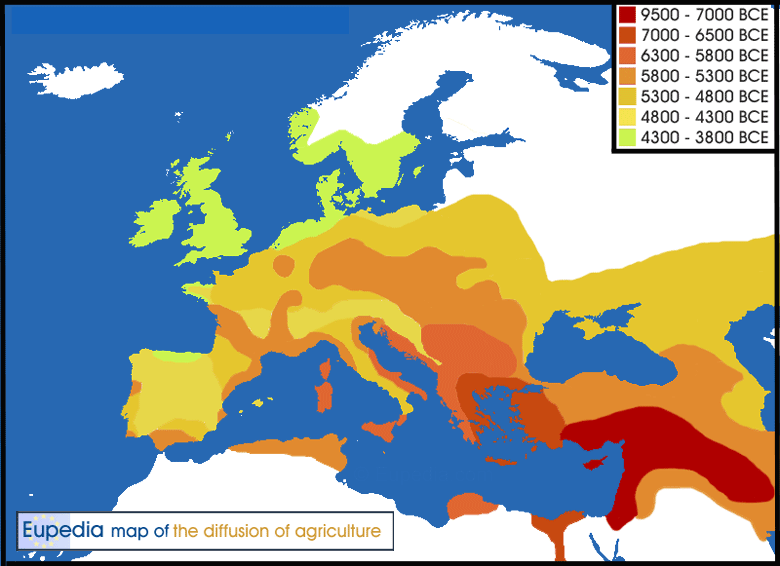
Mathieson et al. (2017) tested numerous individuals from the Neolithic and Chalcolithic Balkans and found the clades L91 (in MN Bulgaria), P303 (in Middle Neolithic Bulgaria and in the Trypillian culture), Z1903 (in Chalcolithic Bulgaria), L42 (in the Trypillian culture), and PF3359 (in the Varna culture).
Ötzi the Iceman (see famous individuals below), Europe's best preserved natural mummy, who died in the Italian Alps 5,000 years ago, during the Chalcolithic, belonged to haplogroup G2a2a2 (L91), a relatively rare subclade found nowadays in the Middle East, southern Europe (especially Sicily, Sardinia and Corsica) and North Africa. G2a2 (PF3146) is otherwise found at low frequencies all the way from the Levant to Western Europe. In conclusion, European Neolithic farmers would have belonged to G2a2a (PFF3146) and G2a2b (L30) and their subclades.
Nowadays G2a is found mostly in mountainous regions of Europe, for example, in the Apennine mountains (15 to 25%) and Sardinia (12%) in Italy, Cantabria (10%) and Asturias (8%) in northern Spain, Austria (8%), Auvergne (8%) and Provence (7%) in south-east France, Switzerland (7.5%), the mountainous parts of Bohemia (5 to 10%), Romania (6.5%) and Greece (6.5%). The hilly terrain of southern Europe indeed makes it ideally suited for herding goats, which G2a men brought with them during the Early Neolithic period. But the most likely explanation is that mountains provided refuge for G2a tribes after the Proto-Indo-European speakers invaded Europe from the steppes of Russia and Ukraine during the Late Copper Age and the Bronze Age (see history of R1a and R1b).
Steppe people were almost exclusively cattle and horse pastoralists and first settled in flat regions like the Hungarian Plain, the North European Plain and the Baltic region. Even after reaching Western Europe, they favoured relatively low lying regions like the Low Countries, western France and the British Isles, where R1b lineages now exceeds 60%, and in some places 80% of the population. In fact, the highest percentages of G2a today are found in the regions last invaded by R1a and R1b people. Indo-Europeans didn't penetrate into Iberia until 1800 BCE and did not cover the whole peninsula until 1200 BCE, and pockets of G2a survive in particularly isolated areas like the Pyrenees, the Cantabrian mountains, or the arid highlands of La Mancha. The Proto-Italics only crossed the Alps into Italy from 1300 BCE and settled more densely in the north, explaining the north-south gradient in R1b in modern Italy, which is practically the mirror of Neolithic haplogroups like G2a and T1a. Sardinians spoke a non-Indo-European language until the Roman conquest, only 2,000 years ago.
The distribution map of all G2a subclades does not impart just how thoroughly Proto-Indo-Europeans eliminated G2a lineages in the northern half of Europe because Proto-Indo-Europeans also carried one type of G2a that was assimiated around the Pontic Steppe. These G2a lineages that were Indo-Europeanised before the great migrations belonged to deep clades of G2a-L140 such as L13 and Z1816 (see below). Nowadays, the Neolithic clades of G2a are found especially in Anatolia, the southern Balkans, the Apennines, central France, and in the Pyrennees. They only represent a tiny fraction of all the G2a in the northern half of Europe, where the Indo-European G2a clades are dominant.
G2a people may have been among the first humans to have acquired the alleles for fair skin. A hunter-gatherer from northern Spain tested by Olalde et al. 2014 still had dark skinned as recently as 7,000 years ago. In contrast, Early Neolithic farmers from the Balkans and Germany already possessed the alleles for fair skin found in modern Europeans. It is still unclear exactly when and among which haplogroup fair skin arose, but it has been suggested that the new diet brought by cereral agriculture would have caused deficiencies in vitamin D, which was traditionally absorbed from fish and meat among foragers. Mutations for light skin would have been positively selected among Neolithic agriculturalists to stimulate the production of vitamin D from sunlight in order to compensate for the scarcity of meat.
Indo-European branches of G2a
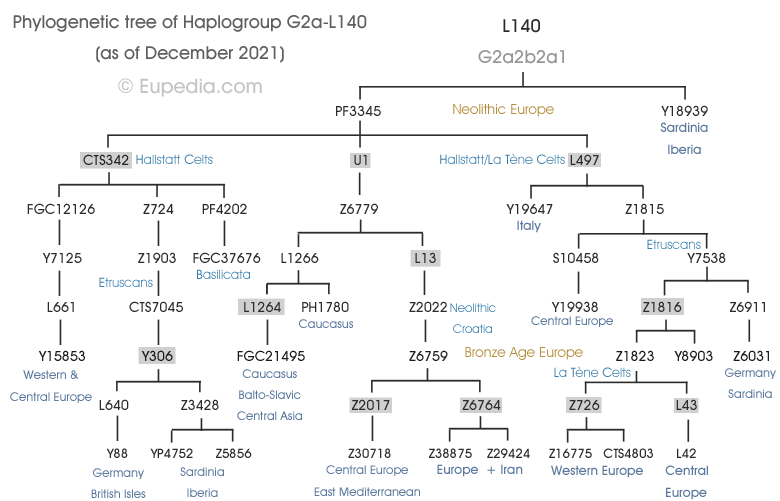
Contrarily to other branches of G2a, which are more prevalent in mountainous areas, some subclades of G2a-L140 are found uniformly throughout Europe, even in Scandinavia and Russia, where Neolithic farmers had only a minor impact. More importantly, G2a-L140 and its subclades are also found in the Caucasus, Central Asia and throughout India, especially among the upper castes, who represent the descendants of the Bronze Age Indo-European invaders. The combined presence of G2a-L140 across Europe and India is a very strong argument in favour of an Indo-European dispersal. However L140 itself emerged over 11,000 years ago, at the onset of the Pre-Pottery Neolithic is far too old to be Indo-European. It is only certain deeper subclades that would have made their way to the Pontic-Caspian Steppe and been absorbed by the Steppe herders before the Yamna period, and would have been redistributed around Europe and Asia by the Indo-European migrations. We should therefore look for subclades that expanded from the Early Bronze Age and are dispersed from northern Europe to Central and South Asia. The best candidates are:
- L1264, which is found in the North Caucasus, in Baltic, Slavic and Germanic countries as well as in Central Asia and India. It was formed 8,000 years ago, but has a TMRCA of only 4,500 years. It would have propagated with haplogroup R1a (Proto-Balto-Slavic and Proto-Indo-Iranian branches).
- L13 came into existence 10,500 years ago, but present carriers all descend from a common ancestors who lived only 5,000 years ago, which corresponds to the Yamna period. Despite its young age, it is found throughout Europe, including Russia, as well as in Central Asia, Iran, the Caucasus, and the Levant. This branch would have spread both with haplogroups R1a and R1b.
- Z1816, which is found throughout Western and Central Europe, and especially in Germanic countries. Its coalescence age was estimated around 4,500 years by Yfull, but its L42 subclade was found in a Trypillian outlier individual who lived between 4,900 and 5,600 years ago. This branch might have been assimilated by Proto-Indo-Europeans through contact with the Late Trypillian people who moved to the Pontic Steppe and merged with Yamna tribes. It would later have spread around Germanic and Celtic countries alongside haplogroup R1b.
The homeland of R1b1a1a2 (M269) and Pre-Proto-Indo-European speakers is presumed to have been situated in eastern Anatolia and/or the North Caucasus. The Caucasus itself is a hotspot of haplogroup G. Therefore, it is entirely conceivable that a minority of Caucasian men belonging to haplogroup G (and perhaps also J2b) integrated the R1b community that crossed the Caucasus and established themselves on the northern and eastern shores of the Black Sea sometime between 7,000 and 4,500 BCE.
An alternative theory is that G2a-L140 came from Anatolia to eastern and Central Europe during the Neolithic (a fact proven by ancient DNA test). Once in Southeast Europe men belonging to the U1 branch founded the Cucuteni-Trypillian culture (with men of other haplogroups, notably I2a1b-L621) around modern Moldova. The Cucuteni-Trypillian people traded actively with the neighbouring with the Steppe cultures, and from 3500 BCE, at the onset of the Yamna period in the Pontic-Caspian Steppe, the Cucuteni-Trypillian people started expanding east into the steppe of what is now western Ukraine, leaving their towns (the largest in the world at the time), and adopting an increasingly nomadic lifestyle like their Yamna neighbours. By the time the Proto-Indo-Europeans started their massive expansion, G2a-U1 men belonging to the L13 and L1264 subclades would have joined R1b and R1a tribes in the invasion of Europe, then of Central and South Asia.
Kura-Araxes branches of G2a
Around the same time as the Indo-European ethnogenesis was taking shape in the Pontic Steppe during the Maykop (3700-3000 BCE) and Yamna (3500-2300 BCE) cultures, another Early Bronze Age society was developping on the other side of the Caucasus: the Kura-Araxes culture (3400-2000 BCE). Although the Kura-Araxes people were less militaristic and more sedentary, they also underwent a major expansion, first west to Anatolia, south to the Fertile Crescent and east toward the Iranian plateau, possibly all the way to Pakistan, where they would have influenced the Indus Valley Civilisation. It is likely that the descendants of the Kura-Araxes culture eventually colonised Greek islands, including Crete, where they would have founded the Minoan Civilisation (2600-1100 BCE), Europe's oldest civilisation. Minoan remains included G2a-P303>pre-L140 (PF3337+, L140-) and J2a1a-L26>Y7010>Y13128>Z36834.
Based on the modern phylogeny, Kura-Araxes people are thought to have belonged primarily to Y-DNA haplogroups J2a1 (the largest lineage), J1a2-Z1828, T1a-P77, G2a1 (L293, aka FGC7535 or Z6552), G2a2b1a (M406) and L1b. Other minor haplogroups may have been present too, including the now rare R1b1a-L388, which was identified in one Kura-Araxes individual by Lazaridis et al. (2016). During the Classical Antiquity ancient Greek islanders, who were descended in great part from the Minoans, colonised southern Italy, bringing their Kura-Araxes lineages with them.
The oldest known G-L293 sample is a Neolithic man from western Iran. Nowadays, G-L293 is the most common G2a clade in the central and northern Caucasus, peaking at 64% of the population in North Ossetia. The Kura-Araxes culture expanded from the South Caucasus, where L293 is considerably lower (1% in Armenia and eastern Turkey). It is nevertheless found at low frequencies across Turkey, northwest Iran, northern Iraq, Syria and Lebanon (i.e. the further extent of the Kura-Araxes culture proper), but also in Bulgaria, Romania, Greece, southern Italy (especially in Sicily) and Corsica. L293 is conspicuously absent from Sardinia and the northern half of Italy, which excludes both a Neolithic and an Indo-European origin.
G-M406 reaches its maximum frequency (18%) among the Chamalal people in Southwestern Dagestan, but is otherwise more common in Georgia (3%), Armenia (2%), Iran (0.5-2.5%) and Turkey (3-6%) than in the Caucasus itself. It is found at low frequencies in the Levant, and as far south as Egypt and Yemen. In Europe, M406 is most common in Greece (2%), including Crete (2.5%), as well as in Italy (3%), and particularly Sicily (4%). Neither G-L293 nor G-M406 have been identified among the hundreds of Neolithic European individuals tested, which is in agreement with a later Bronze Age dispersal from the South Caucasus.
Roman redistribution
By the Iron Age, the G2a population in most of Europe had been decimated by the Indo-European invasions, followed by Celtic warfare. G2a sought refuge from the invaders in the mountains.
The ancient Latins and Romans descend from the Italic tribes who invaded the Italian peninsula from 1200 BCE. They seem to have belonged primarily to haplogroup R1b-U152 (=> see Genetics of the Italian people), but to have carried a substantial minority of G2a-L140 lineages, especially the L13, L1264 and Z1816 subclades. The Latin homeland in central Italy is one of the hotspots for haplogroup G2a in Europe today. The high level in G2a in the Latium might be due to the dual presence of Indo-European L13, L1264 and Z1816 subclades and of earlier Neolithic lineages who descended from the Apennines to live in Rome after being absorbed by the Roman civilisation.
If the ancient Romans and other Romanised peoples from the Italian peninsula had any genetic impact on other parts of the Roman Empire (as they should have), they certainly contributed to a moderate increase of G2a lineages (in addition to R1b-U152 and J2) within the borders of the empire. Indeed, the frequency of haplogroup G decreases with the distance from the boundaries of the empire. Haplogroup G is much rarer in Nordic and Baltic countries nowadays than in Great Britain, despite the fact that agriculture reached those regions around the same time. It is therefore not inconceivable that a part of the G2a in Great Britain, and especially in Wales (where G2a is the highest) should be of Roman origin. Another reason could be that the forested lowlands of northern Germany, Poland and the Baltic were too poor in metals and did not have attract as many Bronze-Age workers from the Caucasus (=> see Metal-mining and stockbreeding explain R1b dominance in Atlantic fringe). Northeast Europe also has a relatively low percentage of haplogroup R1b, which further reinforces the hypothesis that the two haplogroups spread together during the Bronze Age.
Scythian G1
Like G2b, haplogroup G1 (M342) is found across the Middle East as well as in South and Central Asia. During the Neolithic period, whereasa G2a men migrated west across Anatolia to Europe, their G1 cousins migrated east to the Iranian plateau and India. Within Europe, G1 is mostly confined to Romania, Moldova, western Ukraine, eastern Poland, Belarus and Lithuania, with a few samples in central and south Germany. This distribution is reminiscent of R1a-Z93, the Indo-Iranian branch of R1a. Central Asia became a merging zone for southern G1 and J2 lineages with northern R1a lineages during the Bronze and Iron Ages. New hybrid peoples were formed, like the Scythians, who once controlled an empire ranging from northern Pakistan to Xinjiang and to Ukraine. Lineages like R1a-Z93, R1b-Z2103, G1, J2b and Q1b could all have been brought to Europe by the Scythians, Sarmatians and other historical Steppe tribes.
The Romans were known to recruit Scythian or Sarmatian horsemen in their legions. According to C. Scott Littleton in his book From Scythia to Camelot, several Knights of the Round Table were of Scythian origin, and the the legend of Holy Grail itself originated in ancient Scythia. This hypothesis was also taken up in the 2004 movie King Arthur, which opens with the arrival of Scytho-Roman cavalry in Britain. However, Scythians were steppe people would have carried a much higher frequency of R1a-Z93 and R1b-Z2103. Within Britain about a dozen G1 families have been identified in England, and interestingly one family (surname Mills) in Wales, where the Romano-British army of King Arthur supposedly withdrew from the invading Anglo-Saxons. The rare R1b-Z2103 found in Britain were in Dorset and Somerset, including one individual near Glastonbury (surname Allen, which is of Celtic origin, but could also be related to the Alans, a Steppe tribe that spoke a Scytho-Sarmatian language), a place associated with King Arthur.
The maternal lineages (mtDNA) corresponding to haplogroup G
Three of the main maternal lineages thought to have evolved conjointly with Y-haplogroup G2 are mt-haplogroups N1a1a, W1 and X, all minor lineages with roots in the Middle East. Interestingly, N1a, W (aka N2b) and X are directly descended from the very old haplogroup N*, rather than from the more recent macro-haplogroup R (the ancestor of HV, JT and UK, representing 90% of European mtDNA lineages). The long bottleneck evolution of N1a and X mirror that of Y-haplogroup G2. These haplogroups are called Basal Eurasian.
Like G2a none have been found in the Neolithic Levant, but all of them were found in the predominantly G2a populations of Early Neolithic Anatolia and Europe. Of the 40 mtDNA samples from Neolithic Anatolia tested to date, 11 belonged to N1a1a, three to X2 and one to W1. Two others belonged to N1b1a, an even rare lineage today, but closely related to N1a. Nearly half of the mtDNA lineages and slightly over half of Y-DNA lineages in Neolithic Anatolia were therefore Basal Eurasian.
Furthermore, both N1a1a and X2 were found at exceptionally high frequencies in Neolithic Europe compared with present time, N1a1a being found in 13% of samples from the Linear Pottery culture (LBK) and X2 generally ranging between 5% and 10% in various Neolithic cultures. Today, X2 is found in 1.5% of the European population and N1a1a is under 0.5%. The only modern population with high frequencies of mtDNA X are the Druzes, who also have over 10% of Y-haplogroup G. Nevertheless, N1a and X cannot be seen as being exclusively linked to Y-haplogroup G for the simple reason that the first Neolithic farmers from the Fertile Crescent were a compound of several male (and female) lineages, which also included Y-haplogroups E1b1b (at least M123), H2 and T1a (and perhaps minorities of J1 and J2).
Famous individuals

Ötzi the Iceman, Europe's oldest natural human mummy, dating from 5,300 years ago, had his full genome sequenced (the oldest European genome ever tested) and was found to belong to haplogroup G2a-L91 (G2a2a2, formerly known as G2a4).

On 12 September 2012, archeologists from the University of Leicester announced that they had discovered what they believed were the remains of King Richard III of England (1452-1485) within the former Greyfriars Friary Church in the city of Leicester (see Exhumation of Richard III). The skeleton's DNA matched exactly the mitochondiral haplogroup (J1c2c) of modern matrilineal descendants of Anne of York, Richard's elder sister, confirming the identity of the medieval king. Further tests published in December 2014 revealed that his Y-chromosomal haplogroup was G2 (not tested for downstream mutations, but statistically very likely to be G2a3 as a northern European). This however did not match the Y-DNA of three modern relatives (who were all R1b-U152 xL2) descended from Edward III, Richard III's great-great-grand-father. Richard descends from the House of York, while the modern relatives descend from the House of Lancaster via John of Gaunt. Therefore it cannot be determined at present when the non-paternity event occured in the Plantagenet lineage, and whether most of the Plantagenets monarchs belonged to haplogroup G2 or R1b-U152. Both haplogroups are considerably more common in France than in Britain, however, which is consistent with the French roots of the House of Plantagenets.

Joseph Stalin (1878-1953), who ruled the Soviet Union from the mid-1920s until his death, was of Georgian origin and belonged to haplogroup G2a1a. This was determined by testing his grandson, Alexander Burdonsky (his son Vasily's son).

Al Capone (1899-1947), sometimes known by the nickname "Scarface", was an American gangster and businessman who attained notoriety during the Prohibition era as the co-founder and boss of the Chicago Outfit. His seven-year reign as a crime boss ended when he went to prison at the age of 33. According to Geni.com, he was a member of haplogroup G2a-P303.
G2a-Z1816 branch (La Tène Celtic)

Richard Stockton (1730-1781),was an American lawyer, jurist, legislator, and a signer of the Declaration of Independence. His son, another Richard Stockton (1764-1828) was a senator for New Jersey, and his grandson, Robert F. Stockton (1795-1866) was a United States Navy commodore, notable in the capture of California during the MexicanAmerican War. He was a naval innovator and an early advocate for a propeller-driven, steam-powered navy. They descend from Richard Stockton (1665-1709) from Princeton, NJ, whose lineage was determined to be G2a-L497 > Z1816 > FGC809 > Z39863 by the Stockton Surname DNA Project.

The American rock singer Alice Cooper (born Vincent Damon Furnier, 1948), who has been dubbed "The Godfather of Shock Rock", probably belongs to haplogroup G2a-L497 > Z1816 > Z726 > Z16775 > Z17394 > Z34079 (aka FT106898), based on the testing of his distant paternal cousin Vincent Furnier.

Larry Bird (b. 1956), an American professional basketball executive, former coach and former player for the Boston Celtics, is thought to belong to haplogroup G-Z6748 (downstream of Z1816 and Y8903) based on the testing of several relatives descending from Thomas Bird at the Haplogroup G-L497 Y-DNA Project. Larry Bird is the only person in NBA history to be named Most Valuable Player, Coach of the Year, and Executive of the Year.
Other famous members of haplogroup G2a
- Phillip Hamman (1753-1832) : known as "The Savior of Greenbrier", was an American frontier hero who was commended for bravery in the defence of Fort Donnally of Greenbrier County, West Virginia from a Shawnee attack in 1778. Genetic testing of patrilineal descendants of each of Hamman's sons shows his Y-DNA haplogroup to be G2a3b1.
- Najeeb Halaby (1915-2003) : was an American businessman, government official, celebrated aviator, and the father of Queen Noor of Jordan. The Queen was a guest in the PBS television series Faces of America, and one of her paternal cousins had his Y-DNA tested for the purpose of the series. Her paternal lineage was revealed to be haplogroup G.
- Layne Staley (1967-2002) : American musician, songwriter and the original lead singer of the rock band Alice in Chains. He is a patrilineal descendant of Heinrich Stehli (1675-1742) from Switzerland, who is listed as a member of G2a on WikiTree.
- Jake Gyllenhaal : American actor and son of director Stephen Gyllenhaal and screenwriter Naomi Foner. His Y-DNA was revealed by the PBS television series Finding Your Roots.
Read this article in other languages
Follow-up
Ask your questions and discuss about haplogroups on the Forum