Introduction
Who are we? Where do we come from? These are questions that everybody ask at one point or another. All of us learn about our country's history at school. But history has its limits. It won't tell us what makes each of us different from each others.
With the help of science, we can now determine the ancient ethnic origins of one's patrilineal ancestors, by testing the DNA of the paternally inherited Y-chromosome (known as Y-DNA for short).
History books rarely dig into personal genealogies, except for royalty. But even royal families are not immune from non-paternity events. For example, Y-DNA testing revealed that Napoleon III did not carry the same paternal lineage (I2a2a) as his paternal uncle Napoleon I (E1b1b), and was apparently born out of an illicit liaison. But the main purpose of historical population genetics is to trace back one's distant ancestry, going back hundreds, thousands or even tens of thousands of years. The amazing thing with modern genetics is that it is possible to know where our forefathers came from at various periods of history, at least using the patrilineal line. Do we descend from Celts, Slavs, Romans, Germanic tribes, Jews, Phoenicians, ancient Greeks? Until a few decades ago, knowing where one's ancestors lived 2,000 or 3,000 years ago would have been science fiction. It isn't any more.
How can we trace our ancestry with DNA?
We have 23 pairs of chromosomes. The last pair is X-Y for men, and X-X for women. The Y chromosome is the only part of the DNA that does not recombine during procreation. This is because the X and Y chromosomes are of different length, and cannot merge with each others. This explains why the Y chromosome (which we will call "Y-DNA" for short) remains virtually unchanged from generation to generation, and so is practically identical in all men descended from a not-too-distant common ancestor (a few thousand years).
The Y chromosome is a sequence of 59 million characters. Some copying errors (mutations) happen in every generation, like on all the other chromosomes. Each mutation occurring in every new individual is inherited by his descendants. By listing these mutations and by adding up all the mutations found in an individual, it is possible to trace his genealogy and to determine the number of generation that separates it from any other man in the world.
Any man will therefore have the same Y-DNA has his father, brothers, sons, paternal grand-father, and so on, bar a few mutations. This is why all men descending from a same patrilineal ancestor (and therefore having the same surname) share the same series of mutations inherited from all of their accumulated paternal ancestors for thousands of years. All men with the same set of mutations inherited from x thousands of years can therefore be classified in the same family, which population geneticists call a haplogroup. Humanity is thus united in a single large family tree through the Y chromosome, with many branches (haplogroups) and ramifications (subclades), which have evolved over the millennia. The most recent common paternal ancestor to all mankind (also known as Y-chromosomal Adam) lived in Africa at least 300,000 years ago.
Those mutations that occur in every generation are known as Single-Nucleotide Polymorphisms (Single nucleotide polymorphism). They are numbered chronologically based on the time of their discovery.
| More about DNA and SNPs |
|---|
|
Our DNA reads like a book written in an alphabet made of four letters: A, C, G and T. They always go by pairs, A with T, and G with C. Such pairs are called "base pairs", also known as nucleobases. A SNP is a mutation in such a base pair, for example a C replaced by an A. Our genome is divided in 46 chromosomes, which could be seen as volumes of an encyclopaedia. Each chromosomes contains hundreds or thousands of genes, which constitute the chapters. In total there are 3,000 million base pairs. When a mutation occurs, it might alter the expression of a gene, but not always, as some mutations are silent or synonymous. The Y chromosome is made of 59 million nucleobases. As of early 2017, 55,000 SNP's were identified to differentiate the various paternal lineages in the world.
When they are discovered, SNP's get a reference number starting by Rs with a number usually running into the millions. SNP's are used for all the genome, not just the Y chromosome. To assess the genetic risk of develoing a disease, doctors will look at SNP's known to be associated with a particular disease, although hardly any are located on the Y chromosome. Online databases like SNPedia allow people who tested their genome to check for particular variants linked with traits or medical conditions.
To make it less cumbersome for genetic genealogists, DNA testing companies have renamed SNP's with shorter handle names. For example, the mutation rs34276300 defining the Celtic branch of haplogroup R1b, has been renamed P312 by Family Tree DNA. Rivalry between testing companies led them to each use their own nomenclature, so that P312 was named S116 by EthnoAncestry (which became BritainsDNA). What is more some haplogroups are defined by several SNP's (soemtimes hundreds for old haplogroups that underwent prehistoric bottlenecks). At Eupedia you will usually see just the main SNP used on phylogenetic trees to avoid confusion.
=> Read more facts about genetics
|
Population geneticists have classified tens of thousands of Y-DNA mutations commonly found among humans around the world and have rebuilt the genealogical tree of humanity. Human beings experienced severe population bottlenecks during the Last Glacial Maximum (aka LGM, c. 19,000 to 26,000 years ago), particularly in Europe, Central Asia and North Asia, which were partly covered by huge ice caps. Many lineages became extinct during that period. When the population started growing again, men descending from the same tribe carried the same long series of mutations on their Y chromosome, which their last common paternal ancestor had accumulated for several millennia before the population bottleneck occured. Geneticists chose those nodes of sometimes over 100 accumulated common SNP's to define the world's major prehistoric tribes, which they called haplogroups. In other words, people sharing a series of identical unique mutations belong to the same haplogroup, and descend from the same ancestor. It is possible to determine when that ancestor lived based on the number of new mutations that have occurred since then in present-day individuals.
Originally, population geneticists divided humanity into 20 haplogroups, each named by a letter from A to T in chronological order of branching. Haplogroup A represents the source of humanity in Africa. For each new division of a lineage, a number was assigned after the haplogroup. For example, R1 and R2 are two branches of haplogroup R. Numbers and letters then alternate for the successive divisions. For example, R1a and R1b, then R1a1, R1a2, R1b1 and R1b2. Some branches left almost no descendants (e.g., R1a2) while others flourished (e.g. R1a1). The chart below shows when the main haplogroups found in Europe and the Middle East evolved.
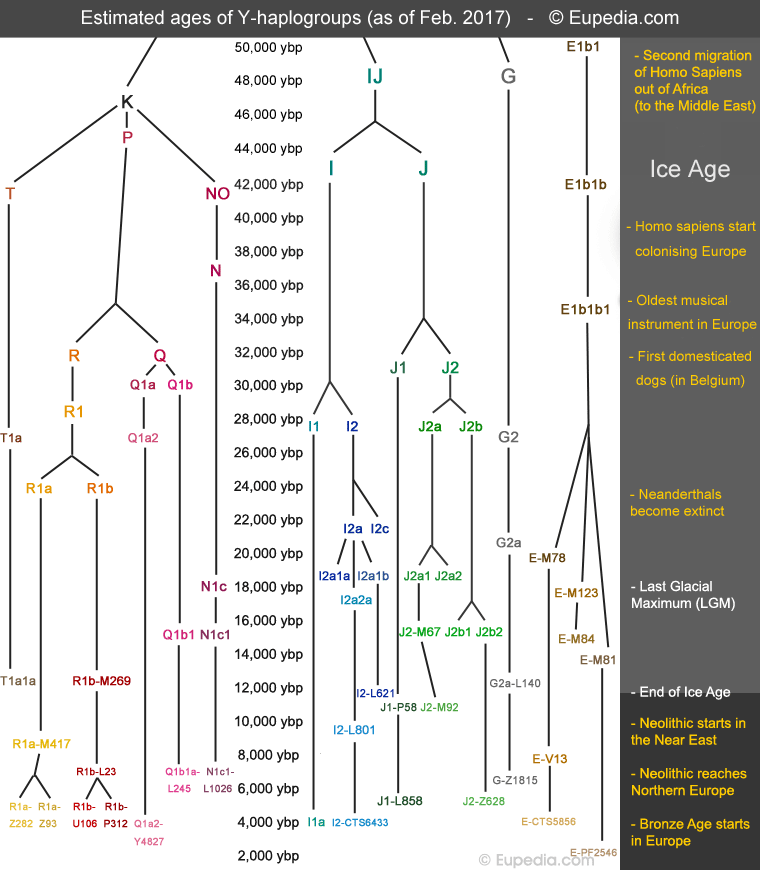
After the Ice Age, humans recolonised the northern half of Europe from LGM refugia in southern Europe. Other tribes moved into Europe from Anatolia and Central Asia. Some 11,000 years ago, agriculture was invented in the Fertile Crescent. A few millennia later, Neolithic farmers spread in all directions, mixing with the Mesolithic hunter-gatherers that were living in Europe and other regions at the time.
5,000 years ago, the first bronze weapons were invented in the North Caucasus by Proto-Indo-European speakers, who had also domesticated horses for the first time in history. Those riders with bronze weapons left the Pontic Steppe of southern Russia and went on to conquer most of Europe, Central Asia and South Asia.
During the Bronze and Iron Ages, the first civilisations arose and expanded. Europe saw the rise and fall of the Celts, the Greeks, the Romans... Then came the great migrations of ancient Germanic, Slavic and Central Asian tribes, followed later by the Vikings.
Each of these migrations spread new genes and new Y-DNA lineages, about which you can learn in detail here, with explanations on the ancient ethnicities linked to each group. You can easily compare the Y-DNA frequencies by country and region and visualise the distribution maps for each haplogroup and their principal subclades.
What about the maternal line?
The same thing can be done on the maternal side using mitochondrial DNA (mtDNA). Mitochondria are organelles that provide energy to cells within the body. They have their own DNA, completely distinct of the nuclear DNA that contains the 23 pairs of chromosomes. This mitochondrial DNA is only passed through mothers as after procreation the spermatozoon loses its mtDNA and the embryo inherits its mtDNA from the mother's ovum.
Although mtDNA was the first genetic method used to trace back ancestry, its scope is more limited because mtDNA is a much shorter sequence (16,569 base pairs) than the Y chromosome, and mutations happen much less frequently than on the Y chromosome. Therefore mtDNA is only useful for tracing back very distant ancestry, typically over 4,000 years ago (read more).
Mitochondria being the cells' powerhouses, mutations in the mtDNA can affect the way the body produces and utilises its energy. Some mtDNA haplogroups have been associated with a more efficient oxygen consumption (VO2 max) and greater physical endurance (e.g. haplogroup H), while others are linked to poorer athletic performance (e.g. J2 and K). Haplogroups U and K were reported to have higher pH in cybrid cells, which confers protection against strokes and neurological disorders and correlate with slightly higher IQ. The C150T mutation, which can potentially be found in any haplogroup, has been linked to increased longevity and resistance to stress. There are many other health-related conditions associated with mtDNA mutations (check the mtDNA haplogroup pages for more details), and that in itself may be a more interesting reason to know one's mtDNA deep clade than for ancestry purposes.
| Want to read more? |
|---|
|
For a more in-depth introduction to historical population genetics, I recommend these two excellent books:
1) Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past by Harvard geneticist David Reich, whose lab pioneered the sequencing of large number of ancient DNA samples. He explains how he and his colleagues tested the whole genome of Neanderthals, discovered by chance a new type of archaic humans, the Denisovans, how much modern humans inherited from archaic humans and how ancient DNA revolutionised our understanding of prehistory, notably by showing that human races 5,000 to 10,000 years ago were radically different from today and that all modern populations are relatively recent blends. The book focuses mostly on how modern ethnic groups came into being and how ancient DNA made it possible to identify these 'ghost populations' from which we descend.
2) Ancestral Journeys: The Peopling of Europe from the First Venturers to the Vikings, by British historian Jean Manco, introduces the subject of historical population genetics then retraces the history of Europe from the Paleolithic to the Middle Ages, including the first farmers, the Copper Age, the Indo-European migrations in the Bronze Age, the Celts and Italics, the Minoans and Mycenaeans, the Etruscans and Romans, the Great Wandering, the Slavs, Bulgars and Magyars, and the Vikings. The book focuses mainly on ancient and modern Y-DNA and mtDNA.
|
How can I test my DNA?
DNA testing is very easy. You just need to order a test kit from a testing company, rub a buccal swab into your mouth (or spit into a small container, depending on the company), and send it back by post. Results typically take from 6 to 12 weeks once the lab has received your sample.
Which DNA test should I choose?
Please check the main article:
Autosomal calculators (GedMatch & Admixture Studio)
Ancient DNA tests have become an incredible tool to elucidate the mysteries of prehistory and ancient migrations. The genomes of hundreds of individuals from the Paleolithic (including Cro-Magnons and Neanderthals) to the Middle Ages (Vikings, Magyars) have been tested, and many of these genomes have been made publicly available. Anyone who has tested his/her autosomal DNA can compare it with any of those ancient samples, be them Neolithic European farmers, Proto-Indo-Europeans from Russia, Iron Age Celts, Roman Britons or Anglo-Saxons, to mention just a few.
A number of autosomal calculators were developed, such as the Dodecad Ancestry Project (by Dienekes Pontikos), Eurogenes (by David Wesolowski), Harappa Ancestry Project (by Zack Ajmal), Fennoscandia Biographic Project (by Anders Pålsen), and Magnus Ducatus Lituaniae Project (MDLP) (by Vadim Verenich and Leon Kull) to compare one's genome with models of historical or regional populations. Some of them use ancient samples as reference populations, so one can get an estimate, for instance, of what percentage of their DNA was inherited from Mesolithic European hunter-gathers vs Neolithic Near Eastern farmers vs Steppe Indo-Europeans. Other calculators attempt to determine the percentage of ancestry linked to Y-DNA haplogroups (e.g. modern R1a distribution in Europe closely matches the East European admixture in Dodecad K12, while R1b resembles West European). Maps are available on Eupedia for several of Dodecad and Eurogenes admixtures, using data from thousands of participants and academic samples for minority ethnic groups (e.g. in the Caucasus).
You have two possibilities to obtain your results from such ancestry calculators:
- Upload your genome's raw data to GEDMatch. In addition to ancestry calculators, this site also looks for relatives among other users. You may not want to use this service if you are concerned about your privacy though.
- Download the Admixture Studio. It's very easy to use. Just unzip the file, run AdmixtureStudio.exe, upload your genome, select the calculator(s) you wish from the list, click 'run' and that's it!
Both of them work with data from all major testing companies (23andMe, Ancestry, FTDNA, Geno 2.0, LivingDNA, MyHeritage).
Companies like MyTrueAncestry also use such calculators to compare your DNA to ancient samples. There is no need to pay for such services. Once you have calculated your admixtures on GEDMatch or Admixture Studio, you can use the free Ancient Ethnicities Analyzer to compare your genome with those of thousands of ancient samples (just like MyTrueAncestry). Furthermore MyTrueAncestry provides misleading results.
If you don't care about autosomal reports provided by testing companies and are only interested in using autosomal calculators or compare your genome on GEDMatch, any autosomal test will do. If you are not interested in your mtDNA results (which are of limited value for ancestry more recent than the Bronze Age) and already tested your Y-DNA or prefer to go with the full Y-DNA sequence (see below), or if you are woman and can't test Y-DNA, you can confidently go with the cheapest autosomal test.
What are surname projects?
Family Tree DNA has thousands of Surname DNA Projects where people can compare their Y-chromosomal DNA with other members with the same or a similar surname and try to determine which members are related and how many generations elapsed since their last common ancestor. To join such a project, you will need to take a specialised STR (Short Tandem Repeats) test, which is different from the SNP test of 23andMe, LivingDNA and Geno 2.0. Surname projects are based on the Y chromosome, since surnames are inherited from one's father and therefore follow the Y-chromosomal lineage. Only men can take this test.
Until recently, the advantage is that these STR tests was that they were more accurate to estimate recent shared ancestry than basic 'backbone' SNP tests. It has consequently become the favoured method for genetic genealogy, especially for people who want to verify their family tree with distant cousins, or ascertain common descent between individuals who share the same surname but lack a paper trail to connect them. However the development of deep SNP tests covering over ten thousands markers, let alone with full Y chromosome tests like Y Elite 2.1 which covers millions of them, has render STR tests somewhat obsolete (and comparatively overpriced). Nevertheless they are still widely used because they were the first type of tests available in genetic genealogy, and surname projects started on Family Tree DNA's website have acquired tens of thousands of participants over the last decade.
Your test will only tell you about your agnatic (patrilineal) line, but nothing prevents you to ask other male family members having a different surname than yours to take a test too. To know your mother's agnatic line, you should test either her father (if still alive), one of her brothers, or one of her paternal uncle (or an uncle's son). The same can be done with your grandmother's agnatic lines, by testing one of her brothers, or male children of a brother. Relatives can even be distant cousins, as long as they are male and have the same surname.
Follow-up
Ask your questions and discuss about haplogroups on the Forum