Quick Facts
- Also known as the Yamnaya Culture, Pit Grave Culture or Ochre Grave Culture.
- Generally considered by linguists as the homeland of the Proto-Indo-European language.
- Probably originated between the Lower Don, the Lower Volga and North Caucasus during the Chalcolithic, around what became the Novotitorovka culture (3300-2700 BCE) within the Yamna culture.
- Highly mobile steppe culture of pastoral nomads relying heavily on cattle (dairy farming). Sheep were also kept for their wool. Hunting, fishing and sporadic agriculture was practiced near rivers.
- First culture (along with Maykop) to make regular use of ox-drawn wheeled carts. Metal artefacts (tools, axes, tanged daggers) were mostly made of copper, with some arsenical bronze. Domesticated horses used as pack animal and ridden to manage cattle herds.
- Coarse, flat-bottomed, egg-shaped pottery decorated with comb stamps and cord impressions.
- The dead were inhumed in pit graves inside kurgans (burial mounds). Bodies were placed in a supine position with bent knees and covered in ochre. Wagons/carts and sacrificed animals (cattle, horse, sheep) were present in graves, a trait typical of later Indo-European cultures.
Maps of the Yamna culture
The Yamna culture and the related Novotitorovska and Maykop cultures

Expansion of the Proto-Indo-European Yamna culture in the Early Bronze Age
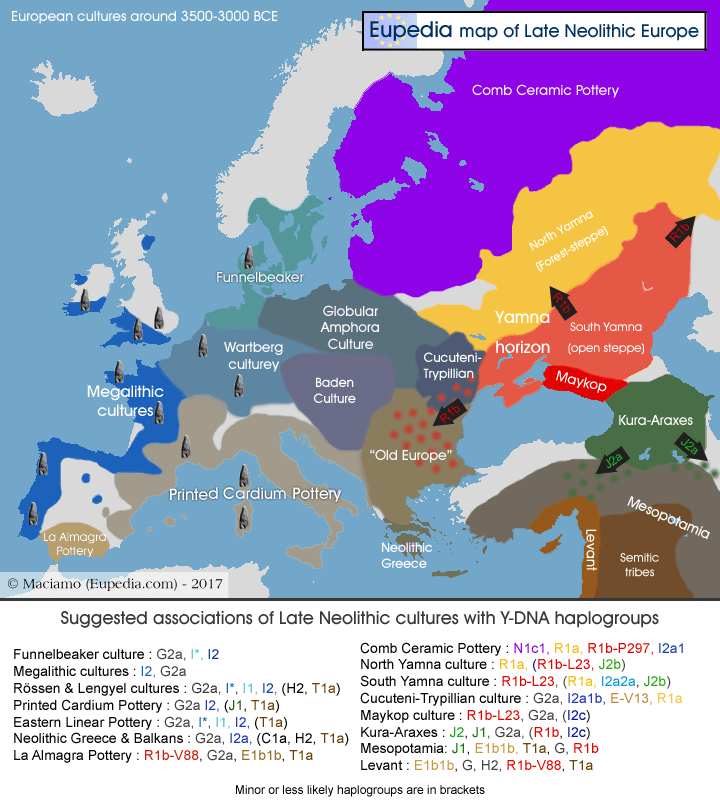
Who were the Yamna people genetically?
Genetic analysis of Yamnayan genomes conducted by Allentoft (2015) and Haak et al. (2015) revealed that Yamnayans were a blend of three ancestral populations. The dominant element (55 to 85%) was the Mesolithic Eastern Hunter-Gatherer (EHG), associated with Y-haplogroups R1a and R1b. Then came the Caucasian Hunter-Gatherer (CHG) admixture, making up about 15-25% of their genomes. This admixture is mainly associated with Y-haplogroup J, and indeed one J* sample was found in Mesolithic Russia. It has been confirmed by ancient DNA that J2b was found among Neolithic farmers from East Transcaucasia (in this case, northwest Iran), and there is convincing evidence that these J2b farmers crossed the Caucasus to settle in the Volga-Ural region during the Early Neolithic. Thirdly, there is an increasing amount of evidence that some R1b tribes crossed from the Steppe to the East Transcaucasia. They would have returned to the Steppe with copper metallurgy during the Khvalynsk culture (5000-3800 BCE), which is ancestral to Yamna. The third admixture was the Western Hunter-Gatherer (WHG), representing about 6% of Yamna genomes, although a few samples lacked it entirely. The WHG admixture is derived from Mesolithic Western Europeans descended from Late Cro-Magnons (Gravettian) and is mainly linked with Y-haplogroup I, and to a lower extent also C1a2 and F.
The Yamna DNA samples recovered from elite Kurgan graves in southern Russia belonged overwhelmingly to haplogroup R1b-Z2103, the essentially eastern branch of Indo-European R1b. The absence of other main R1b subclades is probably due to the dominance of a single royal or aristocratic lineage among the Yamnayan elite buried in Kurgans. Other R1b subclades did show up in Germany (DF27, U152) and Ireland (L21) when Steppe-derived people reached those regions between 2500 and 2000 BCE. The only non-R1b sample found in Yamna was an I2a2a-L699, a lineage descended from WHG tribes that migrated to Eastern Europe, but came back with the Indo-European migrations and is now found especially in Central and Western Europe.
No modern populations possesses a similar genetic admixture as Yamnayans. Among modern Europeans, the CHG admixture found in Yamnayans is most common in the Northwest Europe, but absent from Northeast Europe. Using the Dodecad K12b Gedrosian admixture as a proxy, the highest frequencies are found in the British Isles, (10-13%), Norway, the Low Countries and France (8-10%), then in Germany, Denmark and Sweden (7%), as well as Spain, Portugal and North Italy (5-7%). Haak et al. (2015) compared the genomes of Yamnayans to various modern populations and found that North Europeans (Scots, Scandinavians, Baltics) possessed the highest percentage of Yamna-like ancestry. Since Baltic people (Estonians, Latvians, Lithuanians) lack the CHG/Gedrosia admixture, it does not necessarily mean that they have a lot of Yamna ancestry, but that they descend from a related population with a high percentage of EHG ancestry - more probably actual EHG from Northeast Europe, who only became Indo-Europeanised through the Corded Ware expansion.
On the other hand, ancient DNA confirmed that, circa 2000 BCE, new immigrants to Britain and Ireland who were almost undistinguishable genetically from Unetice people from Central Europe. Modern Irish, Scots and Welsh have the highest percentage of Gedrosian admixture in Europe today and, although Haak et al. only analysed Scots, apparently also the highest percentage of Yamna-like admixture with Norwegians (and presumably Icelanders, who, like the Norwegians, have a relatively high percentage of ancestry from Ireland and Scotland from the Viking Age, especially on the maternal side). The Scots and Irish also happen to have the highest percentage of combined Celto-Germanic R1a (L664 and Z283 subclades) and R1b (P312 and U106), and therefore the highest percentage of patrilineal Yamna ancestry.
In terms of genetic admixtures, the best proxy for ancient Yamnayans would be a person with 3 grand-parents of (Highland) Scottish or Irish descent and 1 grand-parent of Brahui or Balochi descent (both ethnic groups being found in Balochistan, in Southwest Pakistan). Such a person would have about 26% of Gedrosian admixture and 67% of Mesolithic European ancestry. This person would also have a few percents of South Asian and Near Eastern admixture other than the Gedrosian, so it not a perfect match, but as close as it can get using modern populations.
What did Yamnayans look like physically?
According Kurts (1984, p.90), people of the Yamna culture consisted of three distinct phenotypes corresponding to the relatively recent blend between three populations. The dominant type was tall, dolichocephalic, with broad faces of medium height. The second type was more robust with high and wide Proto-Europoid faces. The third type was more gracile, with narrow and high faces of the East Mediterranean type.
The average height for Yamnayan men was 175.5cm (5 ft 9 in), approximately the same as the modern average for American and French men, and slightly taller than the average Mesolithic EHG men, who stood at 173.2 cm.
Yamnayan DNA tested by Haak (2015), Wilde (2014), Mathieson (2015) showed that Yamna people (or at least the few elite samples concerned) had predominantly brown eyes, dark hair, and had a skin colour that was moderately light, lighter than Mesolithic Europeans, but somewhat darker than that of the modern North Europeans. This is not unexpected considering that these samples had about 25% of recent admixture from the Iranian Plateau (before the Indo-European migrations brought Northeast European genes to the region), which would have darkened their pigmentation. Other tests have confirmed that the vast majority of Mesolithic Europeans had blue eyes, and the high incidence of red hair among Northwest Europeans (who have the highest percentage of Yamna ancestry) as well as in the Volga-Ural region and in ancient Chinese depictions of the Tocharians from the Tarim Basin strongly suggest that red hair was found among Yamnayans, and that the genes for red hair (which also include some mutations for fair hair) were spread by R1b Indo-Europeans. (=> see The Origins of Red Hair)
The high CHG admixture in elite Kurgan samples was not found in earlier Steppe cultures, apart from a single R1b sample from the Khvalynsk culture that differed from non-R1b samples in that regard. This indicates that a foreign intrusion from the South Caucasus is responsible for this unusually elevated CHG among the Yamna elite. The considerably lower CHG admixture observed in German Bell Beaker and Unetice samples (average 10%), whih represent the advance of R1b tribes into Central Europe, hints that the rest of the Yamna population had higher Mesolithic European and lower Caucasian ancestry than the Yamna samples tested to date, and could consequently have looked more like modern Scottish and Irish people. This would mean mixed blue and brown eyes, and mixed hair colours with brown hair being predominant, but with a sizeable minority of red hair and perhaps also blond hair. Blond hair appears to have originated among Mesolithic Northeast Europeans, and is therefore be more common in populations with high levels of (Baltic, Slavic and Germanic) R1a.
Y-DNA & mtDNA Analysis
| MtDNA samples from the Yamna culture |
|---|
| Hg |
N1a
|
R0/HV
|
H
|
V
|
J
|
T1
|
T2
|
U2
|
U3
|
U4
|
U5
|
K
|
I
|
W
|
X
|
| N=46 |
1
|
0
|
11
|
0
|
1
|
5
|
7
|
1
|
0
|
3
|
9
|
3
|
1
|
3
|
1
|
| % |
2%
|
0%
|
24%
|
0%
|
2%
|
11%
|
15%
|
2%
|
0%
|
6.5%
|
19.5%
|
6.5%
|
2%
|
6.5%
|
2%
|
The following mtDNA and Y-DNA samples were tested by Haak et al. (2015).
Samples from the Volga-Ural region
| Sample
|
Y-DNA
|
mtDNA
|
Location
|
Date
|
| I0231 |
- |
U4a1 |
Ekaterinovka, Samara |
2910-2875 BCE |
| I0355 |
- |
K1b2a |
? |
? |
| I0357 |
- |
W6 |
Lopatino, Samara |
3090-2910 BCE |
| I0370 |
R1b1a2a2 (Z2103) |
H13a1a1 |
Ishkinovka, Orenburg |
3300-2700 BCE |
| I0429 |
R1b1a2a2 (Z2103) |
T2c1a2 |
Lopatino, Samara |
3339-2917 BCE |
| I0438 |
R1b1a2a2 (Z2103) |
U5a1a1 |
Luzhki, Samara |
3021-2635 BCE |
| I0439 |
R1b1a (P297) |
U5a1a1 |
Lopatino, Samara |
3305-2925 BCE |
| I0441 |
- |
H2b |
Kumanaevskii, Samara |
3010-2622 BCE |
| I0443 |
R1b1a2a (L23) |
W3a1a |
Lopatino, Samara |
3300-2700 BCE |
| I0444 |
R1b1a2a2 (Z2103) |
H6a1b |
Kutuluk, Samara |
3300-2700 BCE |
Most of the following mtDNA samples were tested by Wilde et al. (2014). Deep subclades were not reported in the paper and were determined by Maciamo based on the raw data. The RISE546 to RISE552 samples were tested by Allentoft et al. (2015).
Samples from the Volga region
| Sample
|
Y-DNA
|
mtDNA
|
Location
|
Region/Country
|
| KAL1 |
- |
N1a1a |
Kalinovka I |
Samara Oblast, Russia |
| KAL2 |
- |
H* |
Kalinovka I |
Samara Oblast, Russia |
| NIK1 |
- |
T1a |
Nikolaevka III |
Samara Oblast, Russia |
| NIK7 |
- |
H (rCRS) |
Nikolaevka III |
Samara Oblast, Russia |
| POD1 |
- |
W6 |
Podlesnyj |
Samara Oblast, Russia |
| POD2 |
- |
T2 |
Podlesnyj |
Samara Oblast, Russia |
Samples from the Don and Kuban regions
| Sample
|
Y-DNA
|
mtDNA
|
Location
|
Region/Country
|
| OLE1 |
- |
T2 |
Olennii |
Krasnodar Krai (Sea of Azov), Russia |
| OLE7 |
- |
J2b |
Olennii |
Krasnodar Krai (Sea of Azov), Russia |
| PEJ1 |
- |
U5a1 |
Peschanyi |
Rostov Oblast, Russia |
| RISE240 |
- |
U5a1d1 |
Sukhaya Termista I |
Rostov Oblast, Russia |
| RISE546 |
R1b1a2 (M269) |
U5a1d2b |
Temrta IV |
Rostov Oblast, Russia |
| RISE547 |
R1b1a2a2 (Z2103) |
T2a1a |
Temrta IV |
Rostov Oblast, Russia |
| RISE548 |
R1b1a2a2 (Z2103) |
U4 |
Temrta IV |
Rostov Oblast, Russia |
| RISE550 |
R1b1a2 (M269, xL51) |
U5a1i |
Temrta IV |
Rostov Oblast, Russia |
| RISE552 |
I2a2a1b1b2 (S12195) |
T2a1a |
Peshany V |
Rostov Oblast, Russia |
Samples from central and eastern Ukraine
| Sample
|
Y-DNA
|
mtDNA
|
Location
|
Region/Country
|
| I2105 |
- |
T1a1 |
Shevchenko |
Ukraine |
| I3141 |
- |
H15b1 |
Shevchenko |
Ukraine |
| PES7 |
- |
H1a1 or H5a1j |
Pestchanka II |
Central eastern Ukraine |
| SUG2 |
- |
I1a |
Kirovograd Sugokleya |
Central Ukraine |
| SUG6 |
- |
H1, H3 or H6 |
Kirovograd Sugokleya |
Central Ukraine |
| SUG7 |
- |
H (rCRS) |
Kirovograd Sugokleya |
Central Ukraine |
| SUG8 |
- |
H (rCRS) |
Kirovograd Sugokleya |
Central Ukraine |
| VIN2 |
- |
T1a |
Vinogradnoe |
Southern central Ukraine |
| VIN5 |
- |
T1a |
Vinogradnoe |
Southern central Ukraine |
| VIN12 |
- |
T1 |
Vinogradnoe |
Southern central Ukraine |
Samples from western Ukraine & Moldova
| Sample
|
Y-DNA
|
mtDNA
|
Location
|
Region/Country
|
| MAJ3 |
- |
U5a1 |
Mayaki |
Southwest Ukraine |
| MAJ4 |
- |
U5b2 |
Mayaki |
Southwest Ukraine |
| MAJ5 |
- |
X2h (?) |
Mayaki |
Southwest Ukraine |
| TET2 |
- |
U4a1 |
Tetcani |
Northern Moldova |
Samples from Bulgaria
| Sample
|
Y-DNA
|
mtDNA
|
Location
|
Region/Country
|
| OVI2 |
- |
K |
Ovchartsi |
Southeast Bulgaria |
| OVI3 |
- |
U/K |
Ovchartsi |
Southeast Bulgaria |
| POP1 |
- |
T2a1b |
Popovo |
Southeast Bulgaria |
| POP2 |
- |
U2e |
Popovo |
Southeast Bulgaria |
| POP3 |
- |
U5a1 |
Popovo |
Southeast Bulgaria |
| RIL |
- |
K1 |
Riltsi |
North-east Bulgaria |
Dodecad K12b admixtures of Yamna pastoralists
Y-chromosomal DNA and mitochondrial DNA are useful tools to follow prehistoric population migrations. However, as uniparental markers, they have their limitations and do not inform us about the whole genome composition of individuals, which can evolve very differently due to the randomness of chromosomal recombination, natural selection for specific genes, and of course to the fact that some men can have more children with multiple women, especially in the context of highly unequal prehistoric societies. The Dodecad K12b admixture calculator used here gives estimates of the ultimate region of origin of chromosomal segments outside of the X and Y chromosomes. This page provides a description of each K12b component with a distribution map among the present-day population. Average values for various ancient European populations can be found here.
| Sample ID |
Gedrosia |
Siberia |
NW Africa |
SE Asia |
Atlantic Med |
North Europe |
South Asia |
East Africa |
SW Asia |
East Asia |
Caucasus |
Sub-Saharan Africa |
| I0231 |
27.21 |
1.78 |
0 |
0 |
3.51 |
62.56 |
1.64 |
0 |
0 |
0 |
3.15 |
0.17 |
| I0357_SVP5 |
31.17 |
1.18 |
0 |
0 |
3.08 |
55.89 |
2.47 |
0 |
0 |
0 |
6.21 |
0 |
| I0370_SVP10 |
24.30 |
6.53 |
0.00 |
0.00 |
0.00 |
59.51 |
0.00 |
0.00 |
0.00 |
0.00 |
9.15 |
0.51 |
| I0429_SVP38 |
26.41 |
3.99 |
0 |
0 |
3.2 |
64.51 |
1.24 |
0 |
0 |
0 |
0 |
0.65 |
| I0438_SVP50 |
23.53 |
4.79 |
0 |
0 |
0 |
60.56 |
1.18 |
0 |
0 |
0 |
9.61 |
0.33 |
| I0439_SVP52 |
24.47 |
0.31 |
0 |
0 |
14.14 |
52.37 |
0 |
0 |
0 |
0 |
8.71 |
0 |
| I0441_SVP54 |
31.70 |
0.00 |
0.00 |
1.34 |
0.00 |
56.80 |
10.17 |
0.00 |
0.00 |
0.00 |
0.00 |
0.00 |
| I0443_SVP57 |
27.75 |
2.45 |
0.00 |
0.34 |
5.62 |
59.4 |
0.45 |
0 |
0 |
0 |
2.9 |
1.09 |
| I0444_SVP58 |
30.91 |
0.96 |
0 |
0 |
4.06 |
58.29 |
0.27 |
0 |
0 |
0 |
3.3 |
2.22 |
| I1917 |
25.57 |
2.46 |
0.00 |
0.00 |
8.35 |
36.23 |
0.00 |
0.00 |
0.00 |
0.00 |
26.10 |
1.29 |
| I2105 |
25.51 |
2.91 |
0 |
0 |
2.31 |
59.02 |
1.42 |
0.63 |
0 |
0 |
7.57 |
0.64 |
| I3141 |
24.88 |
3.51 |
0 |
0 |
2.65 |
64.42 |
2.26 |
0 |
0 |
0 |
2.26 |
0 |
| RISE552 |
32.19 |
1.43 |
0.00 |
0.08 |
1.48 |
60.13 |
0.22 |
0.00 |
0.00 |
0.00 |
3.61 |
0.86 |
| RK1001 |
29.25 |
3.69 |
0.00 |
0.00 |
4.17 |
55.21 |
1.32 |
0.00 |
0.00 |
0.00 |
4.74 |
1.63 |
| RK1007 |
29.12 |
2.38 |
0.00 |
0.00 |
0.00 |
53.66 |
5.51 |
0.00 |
0.00 |
0.00 |
8.72 |
0.61 |
| ZO2002 |
24.68 |
1.61 |
0.00 |
0.12 |
1.44 |
56.77 |
2.24 |
0.00 |
0.00 |
0.00 |
12.30 |
0.85 |
|
| Yamna culture |
27.42 |
2.50 |
0.00 |
0.12 |
3.38 |
57.21 |
1.90 |
0.04 |
0.00 |
0.00 |
6.77 |
0.68 |
Read this article in other languages
Follow-up
Find out the latest studies and discuss them on the Ancient DNA Forum.