Geographic distribution
Haplogroup H is the most common and most diverse maternal lineage in Europe, in most of the Near East and in the Caucasus region. The Saami of Lapland are the only ethnic group in Europe who have low percentages of haplogroup H, varying from 0% to 7%. The frequency of haplogroup H in Europe usually ranges between 40% and 50%. The lowest frequencies are observed in Cyprus (31%), Finland (36%), Iceland (38%) as well as Belarus, Ukraine, Romania and Hungary (all 39%). The only region where H exceeds 50% of the population are Asturias (54%) and Galicia (58%) in northern Spain, and Wales (60%).
Haplogroup H possesses approximately 90 basal subclades identified to date, most of which further subdivided in other subclades. The most common subclades are H1, H2, H3, H4, H5, H6, H7, H10, H11, H13, H14 and H20. The Cambridge Reference Sequence (CRS), the human mitochondrial sequence to which all other sequences are compared, belongs to haplogroup H2a2a.
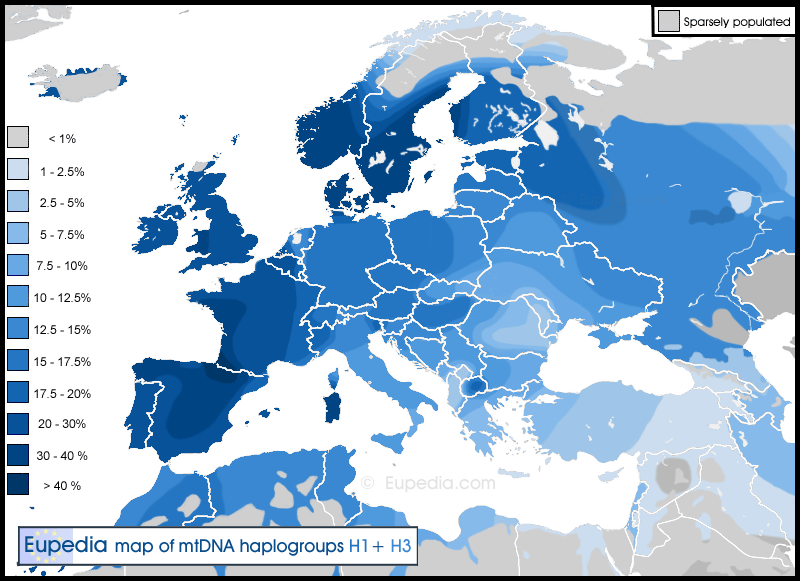
H1 & H3
Haplogroup H1 is by far the most common subclade in Europe, representing approximately than half of the H lineages in Western Europe. Roostalu et al. (2006) estimate that H1 arose around 22,500 years ago. H1 is divided in 65 basal subclades. The largest, H1c, has over 20 more basal subclades of its own, most with deeper ramifications. H1 is found throughout Europe, North Africa, the Levant, Anatolia, the Caucasus, and as far as Central Asia and Siberia. The highest frequencies of H1 are observed in the Iberian peninsula, south-west France and Sardinia. H3 has a very similar distribution to H1, but more confined to Europe and the Maghreb, and is generally two to three times less common than H1.
Distribution of mtDNA haplogroups H1 & H3 in Europe, North Africa and the Middle East
H5
Haplogroup H5 is also found throughout western Eurasia and North Africa, but has a more Central European focus. Its highest frequencies are observed in Wales (8.5%), Slovenia (8%), Latvia (7%), Belgium (7%), Romania (6%), Bosnia-Herzegovina (5.5%), Russia (5%), Germany (5%), Slovakia (5%), Switzerland (4.5%) and Poland (4.5%).
Distribution of mtDNA haplogroup H5 in Europe, North Africa and the Middle East

Origins & History
The mutation defining haplogroup H took place at least 25,000 years ago, and perhaps closer to 30,000 years ago. Its place of origin is unknown, but it was probably somewhere around the northeastern Mediterraean (Balkans, Anatolia or Levant), possibly even in Italy.
Post-LGM recolonisation of Europe
During the Last Glacial Maximum (LGM, c. 26,500 to 19,000 ybp), human populations in Europe were mostly confined to refugia in southern France, Iberia, Italy, the Balkans and the Pontic Steppe. When the climate started warming up and the ice caps progressively receded, people recolonised Europe from these LGM refugia. Only a few ancient mtDNA sequences from this period have been tested. While those from Central and Eastern Europe consistently belonged to haplogroup U (U2, U5, U8), some samples from Spain, Portugal and Italy dating from 18,000 to 10,500 years ago could have belonged to haplogroup H, and possibly even H1 or H3 (although the partial results cannot confirm this with 100% certainty). The modern distributions of H1 and H3 support a Late Glacial recolonisation of Western and Central Europe from the Franco-Cantabrian refugium.
Some people have argued that H1 and H3 only came with Neolithic farmers from the Near East. The oldest incontrovertible evidence of the presence of H1 and H3 in Europe comes from the 5,000-year-old site of Treilles in Languedoc, France. 17 mtDNA sequences were retrieved from this Cardium Pottery culture site, including three that belonged to haplogroup H1 and three to H3. The men belonged to the Near Eastern Y-DNA haplogroup G2a and the Mesolithic European haplogroup I2a. It is noteworthy though that the I2a men belonged exclusively to H1 and H3, a potent, albeit not conclusive evidence that H1 and H3 were lineages that belonged to Mesolithic hunter-gatherers from south-west Europe.
H11 is found across most of northern, central and eastern Europe, but also in Central Asia, where it might have been propagated by the Indo-European migrations (see below). H11a was identified in a Mesolithic hunter-gatherer from the Narva culture in Lithuania by Mittnik et al. (2017).
Other H subclades were also probably found among Mesolithic or Late Upper Paleolithic Europeans based on their exclusive presence in Europe today. This could be the case of haplogroups H4, H10, H17, H45, as well as many minor subclades for which too little data is available at the moment, but that seem to be exclusively European. H4 was found in Neolithic Spain and is found today among both the Basques and the Sardinians, two populations with a high percentage of mixed Mesolithic and Neolithic European ancestry. However H4 was never found among the early Neolithic farmers from the Near East or South-East of Europe. H10 have a stronger presence in Eastern and Central Europe and would have re-expanded from the northern Black Sea region rather than from Southwest Europe after the LGM.
Neolithic diffusion of agriculture
The development of the Neolithic lifestyle in the Fertile Crescent was about to bring a wave of Near Eastern immigrants that would change dramatically the genetic landscape of Europe. Goats and sheep were the first animals domesticated, in the Taurus and Zagros mountains some 11,000 years ago. The domestication of wild cereals could have taken place as early as 23,000 years before present in the southern Levant, although the actual cultivation of wheat and barley didn't start until 11,500 ybp. The first pottery produced in the Fertile Crescent appeared circa 6500 BCE. It is from this period that early agriculturalists began expanding towards western Anatolia and Greece. These Neolithic farmers potentially belonged to haplogroups H5, as well as J1c, K1a, N1a, T2 and X2, all of which have been found in ancient Neolithic samples from Europe and Anatolia, and are also found throughout the Middle East today.
Another Neolithic expansion started off from the Balkans, commencing in Thessaly circa 6500 BCE and spreading at first (6300-5800 BCE) to modern Albania, Macedonia, Bulgaria and Serbia. It is likely that this Thessalian Neolithic was conducted not only by Near Eastern immigrants (Y-haplogroups G2a, and to a lower extent C1a2, H2 and T1a), but also increasingly by local hunter-gatherers assimilated in Southeast Europe (Y-haplogroups E-M78, I*, I1, I2a1, I2a2, I2c). From 5500 to 4500 BCE, the Linear Pottery culture (LBK) spread along the Danube from northern Serbia to Germany and all the way to the Netherlands and Poland. These farmers might also have taken brides among the neighbouring tribes of hunter-gatherers, as suggested by the small amount of Mesolithic European admixture in a 7,500-year old individual from the LBK culture.
Many answers came from the several ancient DNA studies conducted in 2015. Mathieson et al. (2015) tested the genomes of 26 Early Neolithic farmers (6500-6200 BCE) from north-western Anatolia, but only found two members of haplogroup H, both belonging to the H5 lineage - a meagre 7.5% of all lineages. Halogroup H was equally sparse among the oldest Neolithic samples from Southeast Europe. Hofmanová et al. (2015) tested five more Neolithic farmers from the same period, two from the same site in Turkey and three from Greece, but none belonged to H. Szécsényi-Nagy et al. (2015) sequenced 44 mitochondrial DNA profiles from Starčevo culture in Hungary and Croatia, and only three belonged to H (one H5 and two unclear subclades). In other words Early European Farmers only carried about 7% of haplogroup H, against 40 to 50% in present-day Europe. And at present it seems that only H5 indubitably came with early farmers from the Fertile Crescent. The oldest H2, H7 and H20 samples in Europe all date from the Middle to Late Neolithic, and might have come from another wave of settlers from the Middle East, or represent lineages descended from Mesolithic Southern Europeans.
Over 100 Neolithic mtDNA samples from Central, Western and Northern Europe have been tested to date. In addition to the ubiquitous H5, these included H1, H2, H3, H7, H10, H11a, H16, H20 and H89. It is likely that most of them came from assimilated Mesolithic European populations. Haplogroups H1, H10 and H11a in particular are far more frequent in Western Europe (Portugal Spain, France), where paternal lineages also show a much higher percentages of indigenous Mesolithic lineages (I2a1 and I2a2). While it cannot be excluded that subclades like H7 or H20, which are more common in the Middle East than in Europe today, also came with Near Eastern farmers, the majority of H subclades are most likely native to Southern Europe, or in some cases Pontic Steppe and the Caucasus region (H2a1, H6, H8, H13, H15).
H7 was probably part of the Neolithic migration from the Carpathians to Ukraine that gave rise to the Dnieper-Donets culture, along perhaps with mt-haplogroups K1c, K2b, T1a1a, T2a1b1 and T2b, and with Y-haplogroups G2a3b1 and J2b2. All these lineages would subsequently be absorbed by the Proto-Indo-European speakers (Y-haplogroups R1a and R1b) of the Yamna culture during Bronze Age. Nevertheless, H7 could have been native to Southeast Europe and just be among the early maternal lineages to be "Neolithized" by the wave of Near Eastern migrants.
Jones et al. (2015) reported an H13c from Mesolithic Georgia (c. 9650 BCE), indicating that H13 has its roots in the Caucasus. Indeed, nowadays H13 is one of the most common H subclades in the Caucasus (13% in Georgia, 15% in Daghestan), but also in Sardinia (9%), two regions with high levels of Y-haplogroup G2a, J1 and J2a. H13 is also relatively common (2 to 8%) all the way from the Levant to Iberia, but rare in northern Europe. H13 could have been a lineage of goat herders living in the mountainous environment of eastern Anatolia and the Caucasus, then spread through the equally mountainous landscapes of the Mediterranean, from Greece to Italy, then to Iberia.
Neolithic & Chalcolithic Europe
H1 and H3 lineages would have been some of the most prevalent mt-haplogroups among the Megalithic cultures of Western Europe, which spanned the whole Neolithic and Chalcolithic periods, from the 5th millennium BCE until the arrival of the Proto-Celts (Y-DNA R1b) from 2200 BCE to 1800 BCE (or up to 1200 BCE in parts of Iberia). Megalithic people would have belonged essentially to Y-haplogroups I2, G2a and E1b1b, with the possible addition of J2 lineages during the Chalcolithic. From the Bronze Age, R1b male lineages replaced a large percentage of Megalithic Y-haplogroups, but female Megalithic lineages survived almost unchanged in frequency. Celtic culture was born from the fusion of Indo-European paternal lineages (R1b) with native Central and Western European maternal lineages (including H1, H3, H10, J1c, K1a, T2, U5 and X2).
The presence of H1 was confirmed in remains from the Late Neolithic Funnel Beaker culture in Scandinavia, which can also be classified as a Megalithic culture.
Margaryan et al 2017 identified an H2 and an H15a samples among the remains from the Shulaveri-Shomu culture (c. 6000-4000 BCE) in the Caucasus. It is in this culture that the earliest domesticated grapes have been found. Shulaveri-Shomu was the predecessor of the Kura-Araxes culture and may also have influenced the Maykop culture (see below).
Indo-European invasions during the Bronze Age
The Bronze Age brought on a new wave of immigrants from the East that would have a tremendous impact on the gene pool and cultural identity of Europe: the Proto-Indo-European (PIE) people. Originating in the Pontic-Caspian Steppe, to the north of the Black Sea and the Caucasus, PIE speakers were the first people to use bronze technologies for military purposes, starting from c. 3700 BCE. They were also the first to domesticate horses, which they supposedly rode to manage their cattle in the extensive steppes. Already living a (semi-)nomadic existence as cattle herders since the Neolithic (=> see R1b history), horse riding and bronze working provided PIE steppe people with the means to invade and conquer practically any part of the world they wished. They started with Europe, invading the Carpathians and the Balkans repeatedly between 4200 and 2800 BCE, before eventually leaving the steppes almost completely and moving further west across Europe, until the Atlantic coast. The northern branch of PIE speakers, associated with the development of Balto-Slavic and Indo-Iranian languages (=> see R1a), invaded Poland, Germany and Scandinavia to the west from 2900 to 2400 BCE, and Siberia and Central Asia to the east, from 2500 to 1800 BCE.
PIE speakers from the Pontic Steppe are known to have possessed quite different maternal lineages from that of Mesolithic and Neolithic Europeans. A few H subclades linked to the R1a populations of Northeast Europe would obviously have been found among Mesolithic Eastern Europeans, like H1b, H1c and H11. Others may have come from the North Caucasus, like H2a1, or from the advance of Neolithic farmers from the Balkans to Northeast Europe, like H7. Others yet would have come from Anatolia when R1b cattle herders crossed the Caucasus, attracted by the vast pastures of the Pontic Steppe. These would have included H5a, H8 and H15.
Haplogroup H2b, H6a1b, H13a1a1a and many other undetermined H subclades (including many probable H1 and H5) turned up among the mtDNA samples from the Yamna culture, which occupied the Pontic-Caspian Steppe during the Early Bronze Age. The Corded Ware culture, which is associated with the expansion of Y-haplogroup R1a from the steppes to Central Europe and Scandinavia, yielded samples belonging to H1ca1, H2a1, H4a1, H5a1, H6a1a and H10e. Ancient DNA from the Catacomb culture, strongly associated with Y-haplogroup R1a, yielded samples belonging to H1, H2a1 and H6. The Unetice culture, which is thought to mark the arrival of R1b in Central Europe (but overlapping with the previous R1a expansion), had individuals belonging to H2a1a3, H3, H4a1a1a2, H7h, H11a, H82a. Haplogroup H5a was found in the Cucuteni-Trypillian culture and most probably entered the Steppe gene pool by intermarriage.
H6 was absent from Europe before the Bronze Age and has such a wide distribution across the continent nowadays that it would likely have been spread both by R1a and R1b branches of the Indo-Europeans. Indeed, H6 was found in ancient remains from most Indo-European Bronze Age cultures, including Yamna (H6a1b), Corded Ware (H6a1a), Unetice (H6a1b, H6a1b3), Poltavka (H6a2), Okunevo (H6a1b), Srubnaya (H6a1a) and Andronovo (H6). Actually H6 was the only H samples identified so far in the Andronovo culture in Central Asia.
Middle Eastern subclades
Some H subclades are rare in Europe and geographically confined mostly to the Middle East. This includes H14 and H18.
Subclades
Note that only the most common subclades are listed below to avoid excessive sprawling. Deep clades were also cut out. You can find the complete tree on PhyloTree.org.
- H1: found in Europe, North Africa, the Near East, Central Asia and North Asia / found in Neolithic Portugal, Spain, France, Germany, Hungary and Poland
- H1a: found throughout Europe / found in Neolithic Spain and southern France
- H1b: found especially in Eastern Europe, Central Asia and North Asia / found in Bell Beaker Germany, and in Bronze Age Latvia and Russia (Fatyanovo culture)
- H1c: found especially in Eastern Europe, Central Asia and North Asia / / found in Neolithic France and Germany, in Bell Beaker Britain, and in Bronze Age Scotland and Latvia
- H1e: found throughout Europe, including among the Basques, and in Iran (Qashqai) / found in Neolithic France and Germany, in Bell Beaker England, France and Germany, and in LBA France
- H1f: found especially in Finland
- H1g: found especially in Germanic countries
- H1h: found in Central Europe, Britain and Finland
- H1i: found in Ireland and Scotland
- H1j: found mostly in Western Europe, including among the Basques / found in Neolithic Germany
- H1n: found especially in Germanic countries and Finland
- H1q: found in Late Neolithic Spain
- H1t: found among the Basques / found in Late Neolithic Spain
- H1u: found in the Kura-Araxes culture (Bronze Age Armenia) / found in Portugal, Italy, France, Belgium, Britain, Ireland, Slovakia, Finland and Turkey
- H1v: found in Iberia
- H1ae: found in northwest Europe
- H1ag: found in northern Europe
- H1as: found in central and eastern Europe
- H1at: found in northwest Europe
- H1au: found in Poland / found in Neolithic Germany
- H1av: found in Britain and among the Basques
- H1ax: found in Late Neolithic Spain
- H1be
- H1bt
- H1bv
- H2: found throughout Europe and in the Caucasus / found in the Shulaveri-Shomu culture
- H2a: found in Neolithic Italy
- H2a1: found mostly in Eastern Europe, the North Caucasus and Central Asia. IE diffusion (R1a) / found in Bell Beaker Germany, and in Bronze Age France, Scotland and Russia (Fatyanovo culture)
- H2a2
- H2a2a: found throughout Europe / found in LBA Scotland and among the Scythians from Hungary
- H2a2b: found in Chalcolithic Poland (Corded Ware culture)
- H2a3: found in Kenya (El Molo) / found in EBA England
- H2a4
- H2a5: found in Norway, Ireland, Slovakia and among the Basques
- H2a5a: found throughout Europe
- H2a5b
- H2b: found in the Yamna, Srubnaya and Unetice cultures and in Bronze Age Serbia. Found among the Angles and Lombards. Found in Europe, Tukrey, Pakistan and India.
- H2c
- H3: found throughout Europe and in the Maghreb / found in Neolithic Portugal, Spain, France, Germany and Scotland, in Bell Beaker Czechia and in MBA Scotland
- H3a: found in north-west Europe
- H3b: found in the British Isles and Catalonia
- H3c: found in Western Europe, including among the Basques / found in Neolithic France
- H3e
- H3g: found in north-west Europe
- H3h: found throughout northern Europe
- H3i: found in Ireland and Scotland
- H3j: found in Italy
- H3k: found in the British Isles and northern Spain
- H3q: found in Neolithic Alsace (LBK)
- H3r
- H3u
- H3v: found especially in Germanic countries
- H3x
- H3z: found in Atlantic Europe
- H3ao
- H3au
- H4: found especially in central and western Europe, but also in the Near East and Caucasus
- H4a: found in the Neolithic Cardium Pottery culture in Portugal and Spain, in Neolithic Scotland and southern France, in Bell Beaker Czechia and England, in EBA Czechia, and in Bronze Age Bulgaria and Latvia
- H4b: found in Iran
- H4c
- H5: found throughout the Near East and Europe / found in almost all Neolithic cultures in Anatolia and Europe
- H5a: found in the Cucuteni-Trypillian culture
- H5a1: found in most of Europe and Siberia / found in Bell Beaker Netherlands, in Bronze Age Bulgaria, in the Tagar culture (Bronze Age Siberia), and in LBA Scotland
- H5a2: found mostly in Eastern Europe
- H5a3: found in Italy and Siberia
- H5a4
- H5a5: found in Greece
- H5a6: fuond in Germany and England
- H5a7: found in Central Europe
- H5b: found in Bronze Age Bulgaria
- H5b1: found in north-west Europe
- H5b2: found in north-west Europe / found in Bronze Age Russia (Fatyanovo culture)
- H5b3
- H5b4
- H5c: found in England, Germany, Poland, Ukraine / found in EBA England and MBA Wales
- H5d: found in Britain
- H5e: found in England, Germany, Serbia, Russia
- H5p: found in Britain
- H5u: found in Neolithic Alsace
- H6: found throughout Europe, in the Near East and Central Asia
- H6a: found in Bell Beaker Czechia
- H6a1: found in Bronze Age Poland
- H6a1a: found in Bell Beaker Netherlands, in Bronze Age Russia (Fatyanovo culture) and in the Hallstatt culture
- H6a1b: found in Bell Beaker England and Czechia, in MBA Scotland and in Iron Age Tian Shan
- H6b
- H6c
- H7: found mostly in the Near East, Caucasus, Iran, Central Asia and Balto-Slavic countries / found in Bronze Age Poland
- H7a : found in Chalcolithic Poland (Corded Ware culture) and in Bronze Age Bulgaria
- H7b: found among the Tian Shan Huns
- H7c
- H7d: found in the Neolithic Baalberge culutre in Germany
- H8
- H8a: found in the Caucasus (Armenia), Syria, Anatolia and Greece
- H8b: found in Early Roman Lebanon
- H8c: found in central and western Europe, Central Asia and the Altai
- H9
- H10: found throughout Europe / found in Neolithic Germany, France and Portugal
- H10a: found in Bronze Age Latvia
- H10b
- H10c
- H10d
- H10e: found in Bell Beaker Germany
- H10f
- H10g
- H10h
- H11
- H11a: found across most of northern, central and eastern Europe and in Central Asia / found in Mesolithic Lithuania, Middle Neolithic Germany, Megalithic Spain and among Iron Age Latins
- H11b: found in Poland, Slovakia, Serbia and England / found in Bronze Age Poland
- H12: found in Italy (Sicily)
- H13: found mostly around the Caucasus, Iran, Anatolia, and Sardinia, but also across along the Mediterranean coasts of Europe
- H13a
- H13a1 : found in Bronze Age Poland
- H13a2: found among the ancient Caucasian Alans and Tian Shan Huns
- H13b
- H13c
- H14: found mostly in the Near East and Caucasus
- H14a: found mostly in the Levant, Kurdistan, Iran, but also in Bulgaria, Greece, Italy, the Czech Republic, Scotland and Ireland / found in Sumerian Syria and in Hellenistic Lebanon
- H14b: found in Iran, Turkey, Germany and France
- H15: found in Scotland, Germany, Poland, Austria, northern Italy, Central Asia (Turkmenistan), Iran and northern India. Probable Indo-European origin (R1b).
- H15a: found mostly in northwestern Europe / found in the Eneolithic Caucasus (Shulaveri-Shomu culture), in Bronze Age Russia (Fatyanovo culture) and in EBA Scotland
- H15b: found in Sweden, Turkey and India
- H16: found primarily in Germanic countries and Poland, but also in India / found in the Neolithic Rössen culutre in Germany
- H16a: found in England, Germany and Poland
- H16b: found mostly in Germanic countries and Poland
- H17: found in northern Europe
- H17a: found in Sweden, Finland, Lithuania and Germany / found in Bell Beaker Czechia
- H17b: found in Denmark and Ireland
- H17c: found in England and Germany
- H18: found in Portugal, France, England, Germany, Norway, Italy, Turkey and the Arabian peninsula
- H19: found in the Caucasus
- H20: found in England, Hungary, Italy, around the Near East and the Caucasus / found in Neolithic Catalonia
- H21: found in the Caucasus
- H23: found in the British Isles, Scandinavia, the Netherlands, Germany, the Czech Republic, Poland and Russia / found in the Neolithic LBK culutre in Germany
- H24: found in Britain, Ireland, Spain, Germany, the Czech Republic, Poland, Denmark and Finland / found in the Funnelbeaker culutre in Sweden
- H25: found in eastern Germany
- H26: found in Austria
- H26a: found in Ireland, Scandinavia, Germany, the Czech Republic, Croatia and Serbia
- H26b: found in Turkey / found in the Neolithic LBKT culutre in Hungary
- H26c: found in Germany, Romania and Russia (Karelia)
- H27: found in central and northern Europe, but also in Central Asia (Turkmenistan)
- H27a: found in Ireland, northern Germany, Sweden, Finland and northern Russia
- H27b
- H27c: found in Scotland
- H27d
- H28: found in Turkey
- H28a: found especially in Finland, but also in Ukraine, Lithuania, Sweden, Germany and France / found in Bronze Age Latvia
- H29: found in England, Germany, central Italy, Syria (Druzes) and Iran
- H30: found in Italy, central Europe, Norway and the British Isles / found in the Neolithic LBK culutre in Germany
- H31: found in especially in Scandinavia and Scotland, but also in England, Poland and Italy
- H32: found in Bronze Age Serbia
- H33: found in England, Germany, Italy, Greece and Lebanon
- H34: found in England and Germany
- H35: found in England and Slovenia
- H36: found in Finland
- H39: found in Ireland, Britain and Norway / found in the early Neolithic Sopot culutre in Hungary
- H40; found in Portugal, Germany and Poland / found in Bell Beaker Poland and in BA Israel
- H41: found in Britain, northern France, Germany, Sweden, Poland and Romania / found in Bronze Age Russia (Fatyanovo culture) and in Hellenistic Lebanon
- H42
- H44: found in Ireland, England, northern France, Ukraine and Bulgaria
- H45
- H45a: found mostly in Finland
- H45b: found in Ireland
- H46: found in Ireland and Germany / found in the Early Neolithic Germany (LBK) and in Bell Beaker Germany
- H47: found in the Britain, Ireland, the Czech Republic, Italy, Romania, Ukraine, Russia and Armenia.
- H48: found in Ireland, England, France, Germany, Denmark, the Netherlands, Poland and Croatia
- H49: found essentially in Germanic countries, but also in central Italy, the Czech Republic, Poland (Silesia), Russia (Samara) and Azerbaijan
- H51
- H52: found in Britain and Russia (Karelia)
- H53: found in Poland
- H53: found in Ireland, Germany and Belarus
- H55: found in Germany, France and Italy
- H56: found in Britain, Germany, Hungary, Romania and Ukraine
- H58: found in England, Norway, Lithuania, Russia and Italy
- H59: found in Britain and Ireland / found in Bronze Age Poland
- H60: found in Germany
- H61: found in Hungary
- H63: found in Sweden and Germany
- H64: found in Norway
- H65: found in Sweden
- H66: found in England / found in Bronze Age Poland
- H67: found in Scotland
- H71: found in Sweden and France
- H72: found in Russia / found in Bell Beaker Germany
- H73: found in Denmark and Germany
- H76: found in the Czech Republic / found in Bronze Age Bulgaria
- H77: found in France
- H76: found in Ireland, England, the Czech Republic and Tunisia
- H80: found in England / found in Bronze Age Serbia
- H81
- H82: found in Norway, Germany and Poland
- H83: found in England and Germany
- H84: found in Italy (Sicily)
- H87: found in England
- H89: found in the Neolithic Rössen culutre in Germany
- H95a: found in Sweden and north-east Italy
Associated medical conditions
Ruiz-Pesini et al. (2000) reported that men belonging to haplogroup H have the lowest risk of asthenozoospermia (reduced sperm motility).
A study by Baudouin et al. (2005) found that members of haplogroup H had increased chance of survival from severe sepsis at 180 days compared with individuals from other haplogroups.
According to Coskun et al. (2004), mutations that suppress mitochondrial transcription and replication within haplogroup H would place them at increased risk of developing Alzheimer's Disease (AD) compared to other major European haplogroups. However this risk is mitigated between subclades. The highest risk is found in the defining mutation of haplogroup H5a (T4336C), which has been associated with late onset AD by Santoro et al. (2005). In contrast, haplogroups H6a1a and H6a1b have been found to be protective against AD by Ridge et al. (2012)
Hendrickson et al. (2008) studied the role played by mitochondrial function in AIDS progression in HIV-1 infected persons. They found that AIDS progression was slower for members of haplogroups H3, I, K, U, W and X. The follow up study found that haplogroup H was strongly associated with increased lipoatrophy following a antiretroviral therapy.
Martínez-Redondo et al. (2010) found that haplogroup H was associated with a higher maximal oxygen uptake (VO2 max) compared to other haplogroups. This would imply that members of haplogroup H have a greater physical endurance during prolonged exercise. A study conducted by Maruszak et al. (2014) analysed the mtDNA of 395 elite Polish athletes (213 endurance athletes and 182 power athletes) and 413 sedentary controls and found that haplogroup H is among the most overrepresented mtDNA types among endurance athletes at the Olympic/World Class level. The authors found that two polymorphisms in particular were associated with achieving the elite performance level. These were 16080G, which defines haplogroup H1b2 only, and 16362C defines haplogroups H1b1 (subclades a, b, c, d and h only), H1c3b, H1bv1, H3ak, H6, H8, H13b, H20a1a. Another study by Murakami et al. (2002) confirmed the role of 16362C in improved training responsiveness. They also reported three other polymorphisms were associated with increased VO2max, including 16298C, which defines the H6a1a8, and 16325C, which defines the H1an2, H5g, H6b2 and H104 subclades.
The common C150T mutation has been found at strikingly higher frequency among Chinese and Italian centenarians and may be advantageous for longevity and resistance to stress according to Chen et al. (2012). C150T defines haplogroups H1e1a6, H1o, H1as1, H1av1 and H6a1a7, but may also be found among other subclades.
The T16189C polymorphism, defining haplogroup H1a6, H1b, H1f, H1g, H1j9, H1k, H1y, H1z, H1aa, H1ab, H1ac, H1ad, H1ap1, H1cc, H1c3b, H2a2a1g, H3av and H96, lowers the rate of mtDNA replication and consequently the number of mtDNA copies, reducing metabolic efficiency. It has been linked to maternally inherited thinness (Parker 2005), thinness at birth (Soini 2012), an increased body mass index (Liou 2007), and increased frequency of type 2 diabetes in the UK (Poulton 2002) and in Asia (Weng 2005 and Park 2008).
Famous individuals

Various matrilineal descendants of Empress Maria Theresa were tested and confirmed to belong to the same haplogroup (H with the mutations 152C, 194T and 263G, which most probably corresponds to the H3s subclade). This lineage's most recent common matrilineal ancestor is Princess Christine Louise of Oettingen-Oettingen (1671-1747), whose matrilineal descendants include Emperor Joseph II, Emperor Leopold II, Emperor Ferdinand I, Frederick William II of Prussia, Tsar Peter II of Russia, Queen Marie-Antoinette, William I of the Netherlands, Victor Emmanuel II of Italy, and Leopold II of Belgium.


Gill et al. (1994) tested the presumptive mitochondrial DNA of Tsaritsa Alexandra Fyodorovna of Russia and compared it to that of Prince Philip, Duke of Edinburgh. Both being matrilineal descendants of Queen Victoria, they shared the same haplogroup H. The mutations reported were 263G, 16111T and 16357C (an unknown subclade of H). However, King et al. (2004) compared that mtDNA to the relic of Grand Duchess Elisabeth, sister of Empress Alexandra, and found that the sequences did not match and that the HVRI mutations were actually 16129A and 16327T, which would also correspond to haplogroup H, but a different subclade, which cannot be determined without a full sequence test. Whichever is correct, the lineage of Queen Victoria can be said to belong to mt-haplogroup H. Their lineage can be traced back to Anne of Bohemia and Hungary (1503-1546), whose maternal-line descendants include a great many European aristocrats, including (chronologically) Emperor Maximilian II, Marie de' Medici, Emperor Ferdinand II, Władysław IV Vasa, Louis XIII of France, Philip IV of Spain, Charles II of England, James II of England, Emperor Leopold I, William III of England, Louis XV of France, Ferdinand VI of Spain, Leopold I of Belgium, Pedro V of Portugal, Luís I of Portugal, Kaiser Wilhelm II, Ferdinand I of Romania, George II of Greece, Alexander of Greece, Paul of Greece, and Carl XVI Gustaf of Sweden.


Toni de-Dios et al. (2020) recovered Jean-Paul Marat's DNA from the blood-stained paper he was annotating when he was assassinated in 1793 in his bathtub. The French Revolutionary journalist and radical republican politician was found to belong to mt-haplogroup H2a2a1f. His mother came from Castres in the Languedoc.

Lucotte et al. (2010) recovered the DNA of Napoleon Bonaparte from beard hair follicules and compared it to that of his mother, Letizia, and his younger sister Caroline. All three shared the same rare 16184T mutation, which places them within haplogroup H15a1b.

Bogdanowicza et al. (2009) tested the Y-chromosomal DNA and mitochondrial DNA of the exhumed remains of the Renaissance astronomer Nicolaus Copernicus. They established that he belonged to mt-haplogroup H27 (defined by the mutations 16129A and 16316G).

The personal genomics company 23andMe revealed that the mitochondrial haplogroup of business magnate and multi-billionaire Warren Buffett (born 1930) is H4a. He is widely considered the most successful investor of the 20th century. He was ranked by Forbes as the richest person in the world in 2008 and has consistently remained in the top 4 since the annual ranking started in 2000.

The merican singer-songwriter, author, actor, and businessman Jimmy Buffett (b. 1946) was found to belong to haplogroup H3 by the personal genomics company 23andMe. He is known for hit songs such as "Come Monday", "Cheeseburger in Paradise" and "Margaritaville".

The American actress Susan Sarandon (b. 1946), who won an Academy Award for Best Actress for her performance in the 1995 film Dead Man Walking, is a member of haplogroup H according to the BBC programme 'Coming Home' about her father's Welsh ancestry.

Dr. Mehmet Oz (b. 1960), a Turkish-American cardiothoracic surgeon, author, and television personality hosting the The Dr. Oz Show, belongs to haplogroup H2a1 according to the PBS TV series Faces of America.
Other famous members of haplogroup H
Read this article in other languages
Follow-up
Ask your questions and discuss about haplogroups on the Forum